METTL3 Affects Spinal Cord Neuronal Apoptosis by Regulating Bcl-2 m6A Modifications After Spinal Cord Injury
Article information
Abstract
Objective
Spinal cord injury (SCI) is a severe type of neurological trauma. N6-methyladenosine (m6A) modification is one of the most common internal modifications of RNA. The role of METTL3, the predominant methylation enzyme of m6A modification, in SCI remains unclear. This study aimed to investigate the role of methyltransferase METTL3 in SCI.
Methods
After establishing the oxygen-glucose deprivation (OGD) model of PC12 cells and rat spinal cord hemisection model, we found that the expression of METTL3 and the overall m6A modification level were significantly increased in neurons. The m6A modification was identified on B-cell lymphoma 2 (Bcl-2) messenger RNA (mRNA) by bioinformatics analysis, and m6A-RNA immunoprecipitation and RNA immunoprecipitation. In addition, METTL3 was blocked by the specific inhibitor STM2457 and gene knockdown, and then apoptosis levels were measured.
Results
In different models, we found that the expression of METTL3 and the overall m6A modification level were significantly increased in neurons. After inducing OGD, inhibition of METTL3 activity or expression increased the mRNA and protein levels of Bcl-2, inhibited neuronal apoptosis, and improved neuronal viability in the spinal cord.
Conclusion
Inhibition of METTL3 activity or expression can inhibit the apoptosis of spinal cord neurons after SCI through the m6A/Bcl-2 signaling pathway.
INTRODUCTION
Spinal cord injury (SCI) is a common type of neurological trauma that leads to motor and sensory deficits below the injured segment. Severe SCI can even cause death or physical, emotional, and economic consequences for the patient, the family, and society [1,2]. SCI can be traumatic or nontraumatic [3]. Traumatic SCI is caused by direct mechanical injuries such as contusion and compression, while infection and impaired circulation are the etiologies of nontraumatic SCI [4,5]. The local edema, ischemia, and hypoxia caused by the injury in the spinal cord and surrounding tissues, are the main causes of neuronal apoptosis and irreversible spinal cord dysfunction [6,7]. Therefore, it is crucial to hinder the progression of SCI to neuronal apoptosis.
Recently, scientists have found that RNA modifications can be an important target for treating cancer. Particularly, they unfolded that changes in N6-methyladenosine (m6A) modification levels can modulate cancer progression [8,9]. m6A modifications are highly enriched on RNA, especially near the stop codon region. They are catalyzed by methyltransferase and removed by demethylase. Reader proteins recognize the m6A methylation site of RNA [10-12]. m6A modifications have important roles in RNA stability and processing in both physiological and pathological statuses such as immune regulation and cancer [13,14]. m6A methyltransferases mainly consist of METTL3 and METTL14 heterodimers and bind to other regulatory factors such as Wilms’ tumor-associated protein (WTAP) to form the methyltransferase complex [15-17]. METTL3 plays a major catalytic role in completing m6A modification on the shared motif RRACH [14,18].
In most cases, METTL3 is an oncogene that promotes the development and progression of several types of cancer [19-21]. For example, a study by Lin et al. [22] found that METTL3 knockdown in human gastric cancer cell lines, AGS and MKN45, activated apoptosis. Sang et al. [23] showed that METTL3 can promote the development of acute myeloid leukemia through the mdm2/p53 signaling pathway. METTL3 plays a major role in neuronal development, and also in the proliferation and differentiation of stem cells [24-27]. Wang et al. [28] found that METTL3 can control messenger RNA (mRNA) stability and splicing of cerebellar development-related genes. Chen et al. [29] found that METTL3 deletion inhibited neuronal development and shifted adult neural stem cells differentiation toward neuroglia. METTL3 deletion also affected the morphological maturation of newborn neurons in the adult brain. However, there are fewer studies on the role of METTL3 on neuronal apoptosis in SCI. It has been previously reported that Bcl-2, a target gene of METTL3, regulates proliferation and inhibits apoptosis in breast cancer cells [30]. In addition, Zhang et al. [31] indicated that METTL3 increased the expression of Bcl-2 through m6A modification, thereby enhancing the viability and migration of non-small cell lung cancer cells. As a major antiapoptotic protein, Bcl-2 is a crucial factor affecting neuronal apoptosis after injury [32-35]. Based on these findings, Bcl-2 may be a key molecule in inhibiting neuronal apoptosis after SCI. However, it is not clear whether METTL3 can mediate m6A methylation of Bcl-2 in SCI. Therefore, we measured the relationship between m6A, METTL3, and Bcl-2 in SCI models to explore the mechanisms by which METTL3 participates in neuronal apoptosis after SCI. Our findings can provide new insights for treating SCI.
MATERIALS AND METHODS
1. Cell Culture, Differentiation, and Establishment of OGD Model
Rat adrenal pheochromocytoma cell line (PC12 cells) was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). PC12 cell line belongs to a class of neurons that have been widely used in in vitro studies of SCI. PC12 cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI; Gibco, Carlsbad, CA, USA) containing 10% horse serum (Gibco, New Zealand origin), 5% fetal bovine serum (FBS; Gibco, Brazil origin), and 1% penicillin/streptomycin (P/S; Gibco) in a complete culture incubator with CO2, 37°C temperature, and 5% humidity. PC12 cells were inoculated on 6-cm dishes coated with poly-L-lysine and cultured for 24 hours. Cell differentiation was promoted with RPMI 1640 medium containing 10% FBS, 50-ng/mL nerve growth factor (NGF) (Sangon Biotech, Shanghai, China), and 1% P/S [36]. The culture was changed every 2 days and oxygen-glucose deprivation (OGD) was induced for day 5. PC12 cells differentiated by NGF were incubated with serum-free Hanks’ balanced salt solution (Gibco) in 1% O2, 5% CO2, and 95% N2 to establish an OGD model and induce ischemia, early cell injury, and apoptosis [37].
2. Cell Treatment and Establishment of Stable Cell Lines
OGD model was established using a novel METTL3 inhibitor, STM2457 (HY-134836, MedChemExpress, Monmouth Junction, NJ, USA) [38] in RPMI 1640 medium containing 10% FBS, 50-ng/mL NGF, and 1% P/S in PC12 cells for 24 hours. After METTL3 inhibition, incubation continued for 12 hours under glucose-free hypoxic conditions.
Stable cell lines were constructed with a lentiviral system. The plasmid used to construct the stable knockout cell lines was based on short hairpin (sh)RNA-METTL3. To construct the Lenti-METTL3 shRNA vector, METTL3 shRNA oligonucleotides were ligated to the LV2N (U6/Puro) vector. PC12 cells were infected with lenti-METTL3 shRNA in RPMI 1640 complete medium containing 5 μg/mL of polybrene (Gene-Pharma, Shanghai, China) for 24 hours and then screened with 4 μg/mL of puromycin for ≥ 5 days. PC12 cells were collected for reverse transcription quantitative polymerase chain reaction (RT-qPCR) and immunoblotting to detect METTL3 knockdown efficiency. The target sequence of METTL3 shRNA was as follows: shMETTL3, GCTACCGTATGGGACGTTAAC.
3. Animal Model
Forty male Sprague-Dawley rats with 8 weeks of age and 200 ± 20-g body weight were purchased from Fujian Medical University Experimental Animal Center, China. The rats were kept in a temperature-controlled room (22°C–24°C), with 45%–55% humidity and 12-hour light-dark cycles. They had free access to autoclaved food and water. Animal care and experimental procedures were approved by the Animal Ethics Committee of Fujian Medical University (No. IACUC FJMU 2022-0473) and were performed according to the National Institute of Health Guide for the Care and Use of Laboratory Animals. Rats were anesthetized with intraperitoneal injection of 1% sodium pentobarbital (20 mg/kg). For constructing a rat spinal cord hemisection model, the spinal colon was marked on the T9 spinous process and the skin was incised until exposing the T9–10 spinous process. The spinal cord was completely exposed by biting the vertebral plate with biting forceps. The spinal cord was cut on one side with ophthalmic scissors centered on the central canal of the spinal cord. The rats were expected to show a transient lower limb retraction response, indicating the successful establishment of the model. In the sham-operated group, the spinal cord was only exposed [39]. The wound was sutured, and penicillin 8 (U/kg/day for 3 days) was given intraperitoneally to prevent infection.
4. Assessment of Locomotor Capacity
The locomotor function of rats with SCI was assessed using the Basso, Beattie, and Bresnahan (BBB) locomotor rating scale, with scores ranging from 0 to 21. A score of 0 represented no significant movement of the hind limb, and a score of 21 represented full movement. The rats were evaluated on days 0, 1, 3, 7, and 14, postoperatively. The rats moved in the field for 5 minutes, and 2 blinded investigators assessed each rat. The hind limb movement score was recorded, and the mean value was obtained [40].
In the inclined plane test (IPT), the rats were placed on a smooth inclined plane. The slope of the plane started from the horizontal position (0°) and increased by 5° each time. The maximum slope at which the rat remained on the plane for 10 seconds was recorded. The measurement was repeated 5 times for each rat and the mean value was calculated [41].
5. Quantitative Real-Time PCR
Total cellular and tissue RNA was extracted using Trizol (15596018, Invitrogen, Thermo Fisher Science, Waltham, MA, USA). Then, 1 μg of RNA was reverse transcribed using a 5X All-In-One RT MasterMix (G490, Applied Biological Materials, Vancouver, Canada) with reaction conditions of 10 minutes at 25°C, 15 minutes at 37°C, and 5 minutes at 85°C for 5 minutes. The expression levels of mRNA were determined using PerfectStart Green qPCR SuperMix (Lot#Q10228, TransGen Biotech, Beijing, China), and 3 replicates were done for each gene, with Tubulin as the control. The relative expression of each gene was analyzed by the 2-ΔΔCT method. The primer sequences were as follows: Tubulin forward, 5´-TGTATGCCAAGCGTGCCTTTG-3´ and reverse, 5´-AGTATTCCTCTCCTTCTTCCTCACC-3´. METTL3 forward, 5´-AGGGTCTGGATTGCGATGTG-3´ and reverse, 5´-TCAATCTTTCGGGTGCCAGG-3´. Bcl-2 forward 5´-GGATGACTTCTCTCGTCGCT-3´ and reverse 5´-GACATCTCCCTGTTGACGCT-3´.
6. Quantification of m6A RNA Methylation Assay
Total RNA was extracted from tissues and cells using the Trizol method. The overall level of m6A in RNA was detected by EpiQuik m6A RNA Methylation Quantification Kit (P-9005, Colorimetric, Epigentek, Farmingdale, NY, USA). Briefly, 200 ng of RNA was added to each well. Thereafter, capture and detection antibodies were separately added. Detection was performed at 450 nm and the relative m6A levels were calculated for each sample.
7. Western Blotting
Cells and tissues were lysed with RIPA lysis buffer (Beyotime, Shanghai, China) containing protease inhibitors. Total protein was extracted and protein concentration was quantified using a BCA assay kit (Boster Biotech, Wuhan, China). The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% skim milk powder for 3 hours at room temperature, and incubated with the following primary antibodies: METTL3 (ab195352, 1:1,000, Abcam, Cambridge, England), Bcl-2 (ab196495, 1:1,000, Abcam), Bax (#14796, 1:1,000, Cell Signal Technology, Danvers, MA, USA), cleaved-caspase3 (#9664, 1:1,000, Cell Signal Technology), caspase3 (#9662, 1:1,000, Cell Signal Technology), and β-actin (AF0003, 1:1,000, Beyotime). The membrane was incubated with primary antibodies for 15 hours at 4°C and then incubated with horseradish peroxidase-labeled secondary antibodies (A0208, A0216, 1:5,000, Beyotime) for 3 hours at room temperature. The protein bands were detected using chemiluminescence reagents (Beyotime). Quantification was performed using ImageJ software.
8. Bioinformatics Analysis
METTL3 expression was analyzed using data from the control and SCI 3-, 7-, and 14-day groups from the GSE45006 dataset (https://www.ncbi.nlm.nih.gov/). m6A modified motif distribution and secondary structure of Bcl-2 mRNA were analyzed using the SRAMP database (http://www.cuilab.cn/sramp). RMBase v2.0 (https://rna.sysu.edu.cn/rmbase/index.php) was used to identify METTL3 binding to its target. The main m6A motifs of METTL3 after binding to the target genes were identified. The binding region of Bcl-2 mRNA to METTL3 was predicted using catRAPID omics v2.0 (http://service.tartaglialab.com/page/catrapid_omics2_group).
9. m6A-RNA Immunoprecipitation Assay and RNA Immunoprecipitation Assay
m6A-RNA immunoprecipitation (meRIP) assay was performed using the RiboCluster Profiler RIP-Assay Kit (RN1001, MBL, Hokkaido, Japan), according to the manufacturer’s protocol. Briefly, the kit can extract RNAs that are coincubated with a complex of magnetic beads and m6A antibody (ab286164, 5 μg, Abcam). The RNA bound to the magnetic beads was eluted and purified. RT-qPCR was used to detect the target gene expression levels.
RNA immunoprecipitation (RIP) assay was performed using the RiboCluster Profiler RIP-Assay Kit (RN1001, MBL) to purify the target genes. IgG and METTL3 antibodies (15073-1-AP, 5 μg, Proteintech, Rosemont, IL, USA) were conjugated to magnetic beads for the enrichment of target genes. RT-qPCR was performed to detect the enrichment level of Bcl-2 RNA.
10. Cell Counting Kit-8 Assay
Cell viability was measured using the Cell Counting Kit-8 (CCK-8) kit (Dojindo Laboratories, Kyushu, Japan) according to the manufacturer’s protocol. Briefly, PC12 cells were inoculated into 96-well plates (1× 10~4 per well). After differentiation, the cells were treated based on their groups. Then, 10 μL of CCK-8 solution was added to each well. After 2 hours of incubation, the absorbance was read at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
11. TUNEL Assay
PC12 cells were fixed and permeabilized. Then, they were incubated with a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) kit (Roche, Basel, Switzerland) for 2 hours at 37°C, and the nuclei were stained with 4´-6-diamidino-2-phenylindole. Images were taken by a fluorescent microscope (Leica, Heidelberg, Germany) and adjusted for exposure intensity. Positive cells were counted using ImageJ software (National Institute Health, Bethesda, MD, USA).
12. Flow Cytometry
The apoptosis rate was detected using Annexin V-FITC/PI assay kit (BD Bioscience, San Jose, CA, USA). Cells were collected and washed twice with phosphate-buffered saline and dark stained for 15 minutes at room temperature. The apoptosis rate was measured by flow cytometry (FACSCalibur, BD bioscience) and analyzed by Flow Jo software (BD, Ashland, OR, USA).
13. Statistical Analysis
All data were analyzed using IBM SPSS Statistics ver. 21.0 (IBM Co., Armonk, NY, USA). Data are presented as mean±standard deviation in this article. Dataset was assessed for normal distribution and homoscedasticity. Comparisons between the 2 groups were performed by t-test, and comparisons between more than 2 groups were performed by 1-way analysis of variance and Tukey test. A p-value of < 0.05 was considered statistically significant. Data from at least 3 independent experiments were collected for each group.
RESULTS
1. OGD Damage Can Increase METTL3 Expression and m6A Modification Levels
As the most prevalent methylation modification on messenger RNA and noncoding RNAs, m6A is involved in RNA stabilization, degradation, and processing, thereby regulating gene expression [42]. Previous studies found that silencing METTL3 reduced m6A methylation levels in OGD/R-induced LNC-D63785 and decreased neuronal apoptosis [43]. However, the role of METTL3 in SCI has not been fully elucidated. Therefore, we aimed to investigate whether METTL3 can influence the outcome of SCI by regulating m6A. We first constructed an OGD model using PC12 cell line and measured METTL3 transcription levels and overall m6A modification levels at different time points of OGD (3, 6, and 12 hours). The results showed that the transcription level of METTL3 and the overall m6A modification level gradually increased after inducing OGD (Fig. 1A, B). Western blotting also indicated the same results. The protein levels of METTL3 also gradually increased after OGD (Fig. 1C). These findings revealed that the change of METTL3 depends on the duration of OGD, and the highest METTL3 expression level was found in the 12-hour group. For all of the subsequent experiments, 12 hours of OGD was used to induce the injury.

Changes in METTL3 expression and m6A modification in PC12 cells after OGD. (A) Reverse transcription quantitative polymerase chain reaction detected the transcript levels of METTL3 at different time points. (B) m6A modification measurement by m6A colorimetric assay at different time points after OGD. (C) Western blotting analysis for METTL3 expression at time points after OGD. m6A, N6-methyladenosine; OGD, oxygen-glucose deprivation. The significance levels are shown as follows: ns, not significant; versus 0-hour group: *p < 0.05, **p < 0.01, ***p < 0.001.
2. SCI Increases METTL3 Expression and m6A Modification in Rats
Recent studies have shown that SCI can change m6A methylation in rats, which is closely related to its prognosis [44]. We measured METTL3 expression and overall m6A modification level in a rat model of spinal cord right hemisection to further explore the role of MEETL3 in SCI. First, we assessed rat movement after SCI using the BBB score and the IPT. After 3, 7, and 14 days of surgery, the BBB score and tilt angle gradually increased, and the mobility of rats gradually recovered from the 3rd day. After 14 days, the right lower limb regained most of its mobility, but all indicators were at a lower level compared with the sham group (Fig. 2A, B). Using the GSE45006 dataset in the National Center for Biotechnology Information database, we selected the METTL3 expression values of the control group and 3 other SCI samples (3, 7, and 14 days) and found that METTL3 expression level was highest on the 3rd day and then gradually decreased (Fig. 2C). The transcription levels and overall m6A modification levels of METTL3 were detected by RT-qPCR and m6A colorimetric assay at different time points (3, 7, and 14 days) after spinal cord hemisection. Similar to the overall m6A modification level, the transcription level of METTL3 was highest on the 3rd day, then gradually decreased (Fig. 2D, E). Western blotting showed that the protein level of METTL3 was also highest on the 3 days and gradually decreased afterward (Fig. 2F). The results demonstrated that decreased METTL3 expression was inversely associated with the recovery of spinal cord function in rats. Therefore, METTL3 expression is closely related to the overall m6A modification level, which reflects the severity of SCI.
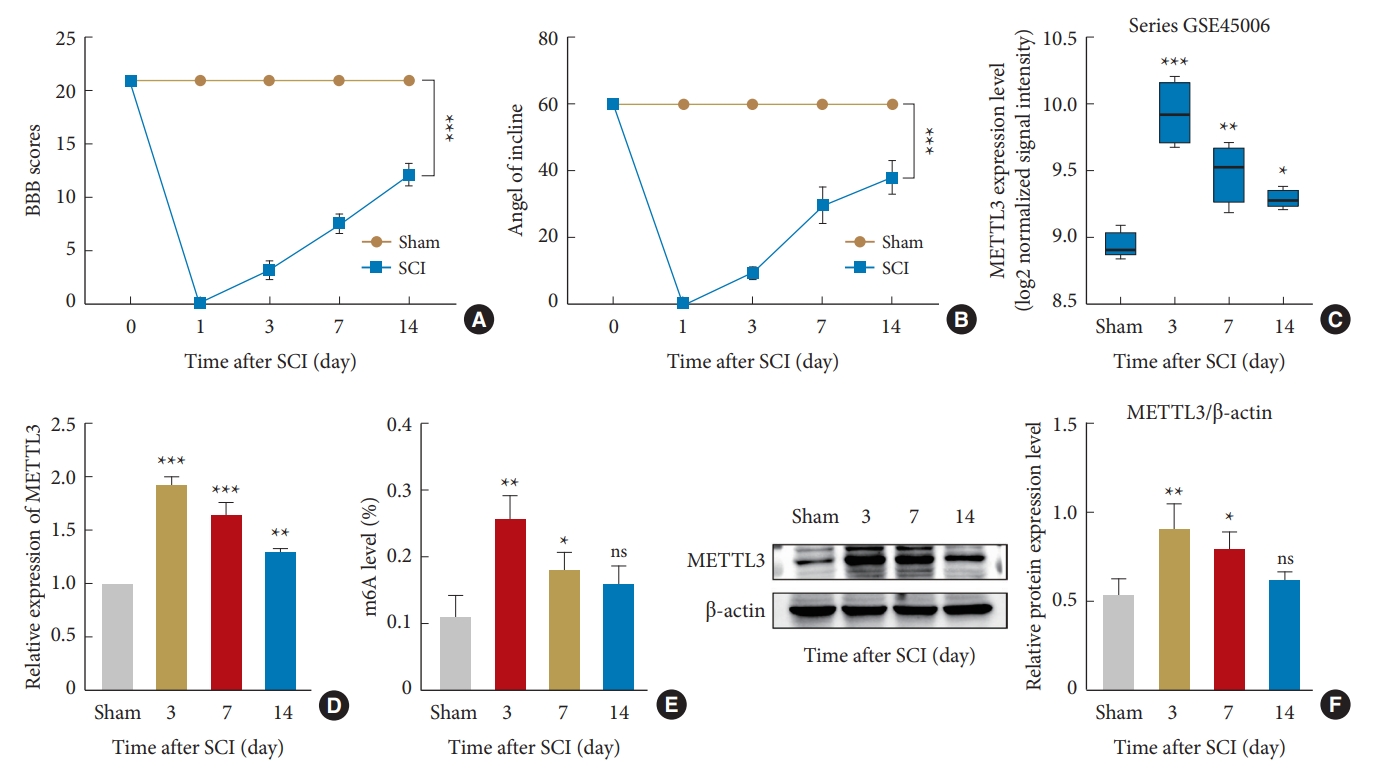
Changes in METTL3 expression and m6A modification after spinal cord hemisection in rats. (A) BBB locomotor rating scale to assess hindlimb motor function. (B) Inclined plane test to observe the changes in tilt angle at different time points after spinal cord injury (SCI). (C) Analysis of METTL3 expression at different time points using the GSE45006 dataset. (D) Reverse transcription quantitative polymerase chain reaction to detect changes at different time points. (E) m6A modification measurement at different time points by m6A colorimetry. (F) Western blotting for measuring the protein expression of METTL3. BBB, Basso, Beattie, and Bresnahan; m6A, N6-methyladenosine. The significance levels are shown as follows: ns, not significant; versus sham group: *p < 0.05, **p < 0.01, ***p < 0.001.
3. Predicting the m6A Site of Bcl-2 mRNA and Its Interaction With METTL3
Previously, it has been shown that Bcl-2 mRNA has m6A modifications, which can influence its expression with METTL3 [30,31,45]. Therefore, we used the SRAMP database to predict the possible presence of m6A methylation modifications in Bcl-2 mRNA and the most likely secondary structure of mRNA (Fig. 3A, B). It has been shown that after completing m6A modification in the nucleus, METTL3 can remain bound to the transcript. After translocation into the cytoplasm, METTL3 can bind to eIF3, which may cause a link between METTL3 at the 3´ UTR of the mRNA and the 5´ mRNA cap to create an mRNA loop. The mRNA loop can enable the ribosome on the termination codon to reload into the transcript and directly activate translation [46]. Understanding this secondary structure can help us to explore other functions of METTL3. We also used the RMBase v2.0 database to predict the m6A motif of Bcl-2 mRNA in the GSE-2460366 dataset and found that the m6A modification was mainly enriched between the coding sequence and 3´-untranslated region, mainly near the termination codon (Fig. 3C). Meanwhile, the interaction of Bcl-2 mRNA with METTL3 was analyzed using the catRAPID database. The results showed a strong binding between the 300th and 600th nucleotides (Fig. 3D). This was different from the m6A-rich position, suggesting that METTL3 itself may have 2 structural domains: a catalytic domain, mainly responsible for m6A modifications, and a binding domain, to anchor on RNA and facilitate m6A modifications. Previous studies have shown that METTL3 consists of a zinc finger domain (ZFD) and a methyltransferase domain, and ZFD is critical for target recognition [18,47,48]. However, it has been reported that METTL3 and METTL14 can form a complex and that METTL14 mainly helps METTL3 bind to RNA [10,13,48]. Therefore, the binding domain of METTL3 may assist the catalytic domain to complete m6A modification in specific regions by anchoring on specific sites. On the other hand, we used RIP and meRIP to verify the relationship between Bcl-2 mRNA and m6A modification. The results of meRIP showed that the m6A-immunoprecipitation group had higher enrichment levels of Bcl-2 mRNA compared with the IgG group. In contrast, the METTL3-IP group possessed a higher Bcl-2 mRNA enrichment level than the IgG group (Fig. 3E, F). In summary, METTL3 interacts with m6A modification on Bcl-2 mRNA, indicating that the m6A modification on Bcl-2 mRNA is catalyzed by METTL3.
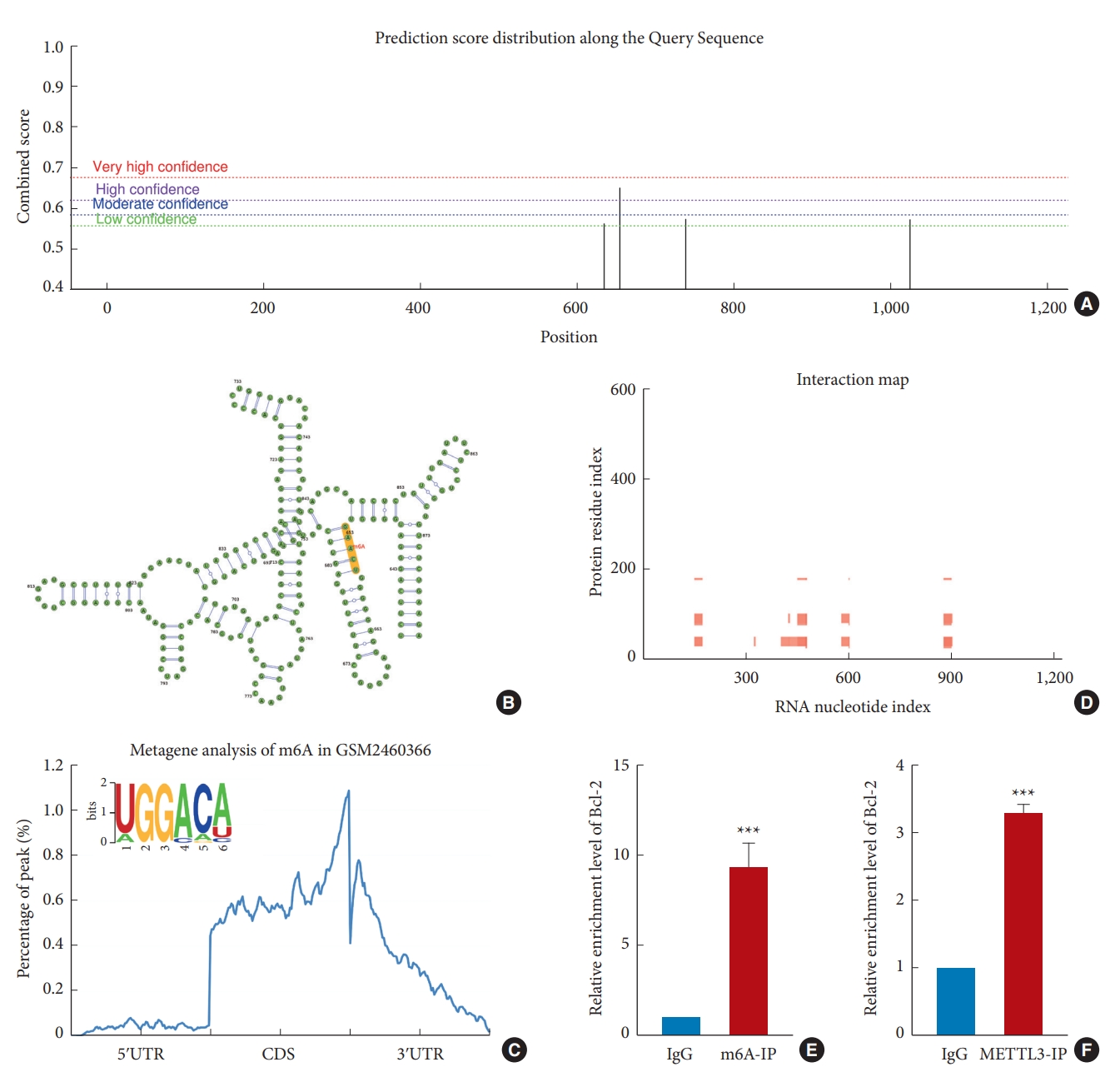
m6A modification sites and common motifs and the interaction between METTL3 with Bcl-2 mRNA. (A) The distribution of m6A modification on Bcl-2 mRNA. (B) The secondary structure of Bcl-2 mRNA. (C) Distribution of m6A modification in Bcl-2 target genes and common motifs (GGACA). (D) The binding site of METTL3 on Bcl-2 mRNA. (E) meRIP shows the presence of m6A modification of Bcl-2 mRNA. (F) RIP shows that METTL3 interacts with Bcl-2 mRNA. Bcl-2, B-cell lymphoma 2; mRNA, messenger RNA; m6A, N6-methyladenosine; meRIP, m6A-RNA immunoprecipitation; RIP, RNA immunoprecipitation; M6A-IP, N6-methyladenosine-immunoprecipitation; METTL3-IP, methyltransferases like 3-immunoprecipitation; 5ʹUTR, 5ʹ-untranslated region; CDS, coding sequence; 3ʹUTR, 3ʹ-untranslated region. The significance levels are shown as follows: versus IgG group: ***p < 0.001.
4. STM2457 Inhibits METTL3 Activity, Alters Bcl-2 Expression, and Regulates Neuronal Apoptosis
STM2457 has excellent inhibitory activity against METTL3 [49-51]. We used the CCK8 method and m6A colorimetric assay to detect the cellular activity and overall m6A modification degree with different drug concentrations (5, 10, 15, 20, 25 μM). The results showed that the cell activity was not affected between 0–25 μM. The overall m6A modification degree gradually decreased with increasing concentrations between 0 μM and 10 μM, while remaining constant between 10 μM and 25 μM (Fig. 4A, B). Therefore, we chose 10 μM as the effective inhibitory concentration for the OGD model. As the major antiapoptotic protein, Bcl-2 can stop programmed cell death [52,53]. Therefore, we used RT-qPCR to detect the transcription levels of Bcl-2 after OGD. There was no difference between the control group and the control+STM2457 group, indicating that the expression level of Bcl-2 mRNA was not affected by 10 μM of STM2457, while it decreased in the OGD group. Compared with the OGD group, the expression level of Bcl-2 mRNA significantly increased in the OGD+STM2457 group, indicating the close link between METTL3 function and Bcl-2 transcription after OGD (Fig. 4C). Then, we used western blotting to detect the expression level of apoptosis-related proteins. The results showed that there was no significant difference between the control group and the control+STM2457 group, but Bax/Bcl-2 and cleaved-caspase3/caspase3 ratios were higher in the OGD group compared with the control group. Furthermore, these ratios were significantly lower in the OGD+STM2457 group compared with the OGD group (Fig. 4D). Next, TUNEL staining was used to measure the apoptosis rate in each group. The number of TUNEL-positive cells significantly increased in the OGD group compared with the control group. The number of TUNEL-positive cells was significantly lower in the OGD+STM2457 group compared with the OGD group (Fig. 4E). We also performed Annexin VFITC/PI flow cytometry to measure apoptosis rate. We found that the total apoptosis rate was significantly higher in the OGD group compared with the control group. In addition, Annexin V-FITC/PI flow cytometry revealed that the apoptosis rate was significantly lower in the OGD+STM2457 group than in the OGD group (Fig. 4F). This shows that METTL3 inhibition can enhance Bcl-2 transcription, increase its protein expression, and inhibit neuronal apoptosis.
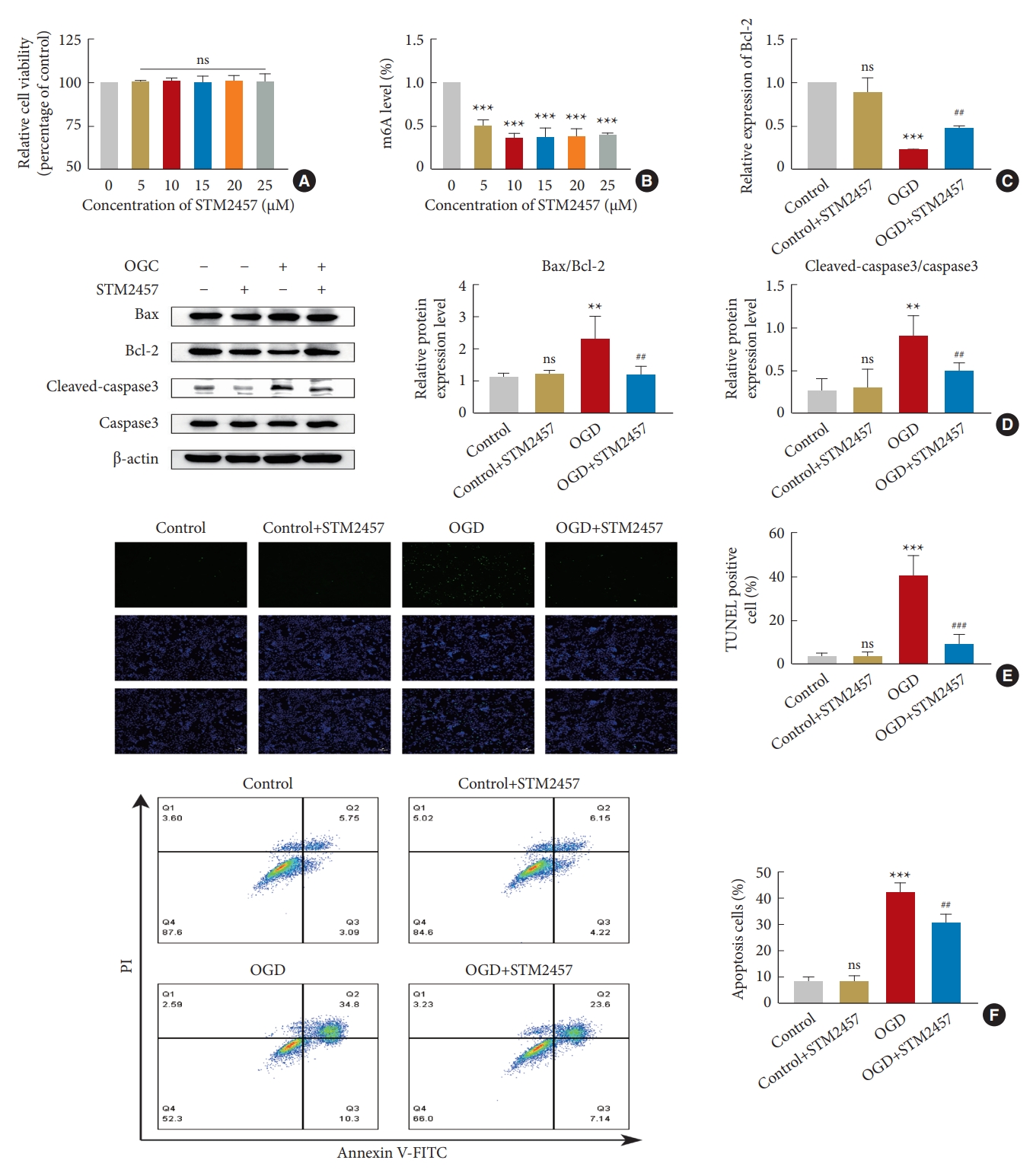
STM2457 regulates neuronal apoptosis by inhibiting METTL3 activity. (A) Cellular activity was not affected at drug concentrations of 0–25 μM. (B) STM2457 decreased the overall m6A modification level. (C) STM2457 enhanced Bcl-2 transcription under OGD. (D) Western blotting of Bax, Bcl-2, cleaved-caspase3, and caspase3 in the presence of STM2457. (E) TUNEL assay of PC12 cells for assessing apoptosis. (F) Flow cytometry for PC12 cell apoptosis. m6A, N6-methyladenosine; Bcl-2, B-cell lymphoma 2; mRNA, messenger RNA; OGD, oxygen-glucose deprivation; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; V-FITC, V-fluorescein isothiocyanate. The significance levels are shown as follows: ns, not significant; versus control group: *p < 0.05, **p < 0.01, ***p < 0.001; #versus OGD+STM2457 group: #p < 0.05, ##p < 0.01, ###p < 0.001.
5. METTL3 Knockdown Increases Bcl-2 Expression and Attenuates OGD-Induced Neuronal Apoptosis
We knocked down METTL3 by lentiviral infection of PC12 cells to investigate whether METTL3 inhibition can inhibit apoptosis by upregulating Bcl-2. After 24 hours, we detected the changes in METTL3 expression level and overall m6A modification. RT-qPCR, western blotting, and m6A colorimetric analysis demonstrated that the mRNA and protein levels of METTL3 and overall m6A modification were significantly lower in the sh-METTL3 group compared with the wild type (WT) and negative control (NC) groups (Fig. 5A-C). RT-qPCR was used to detect the transcription level of Bcl-2 after OGD injury. There was no significant difference between the OGD+WT group and the OGD+NC group, while the transcription level of the OGD+sh-METTL3 group increased (Fig. 5D). Next, we used western blotting to detect the expression levels of apoptosis-related proteins. Compared with other groups, the Bax/Bcl-2 and cleaved-caspase3/caspase3 ratios decreased after METTL3 knockdown (Fig. 5E). Meanwhile, cell activity was detected using the CCK8 method. After OGD, the cell activity was significantly higher in the OGD+sh-METTL3 group than in the other groups (Fig. 5F). The apoptosis rate was detected using TUNEL staining, and it was observed that the number of TUNEL-positive cells significantly reduced in the OGD+sh-METTL3 group and was lower than the number of TUNEL-positive cells in the OGD+WT and OGD+NC groups (Fig. 5G). The apoptosis rate was also measured by Annexin V-FITC/PI flow cytometry. We found that the total apoptosis rate was significantly higher in the OGD+WT and OGD+NC groups and there was not a significant difference between them. The total apoptosis rate was significantly lower in the OGD+sh-METTL3 group than in other groups (Fig. 5H). These results suggest that METTL3 knockdown can decrease m6A levels, increase the mRNA and protein levels of Bcl-2, and inhibit neuronal apoptosis after OGD.
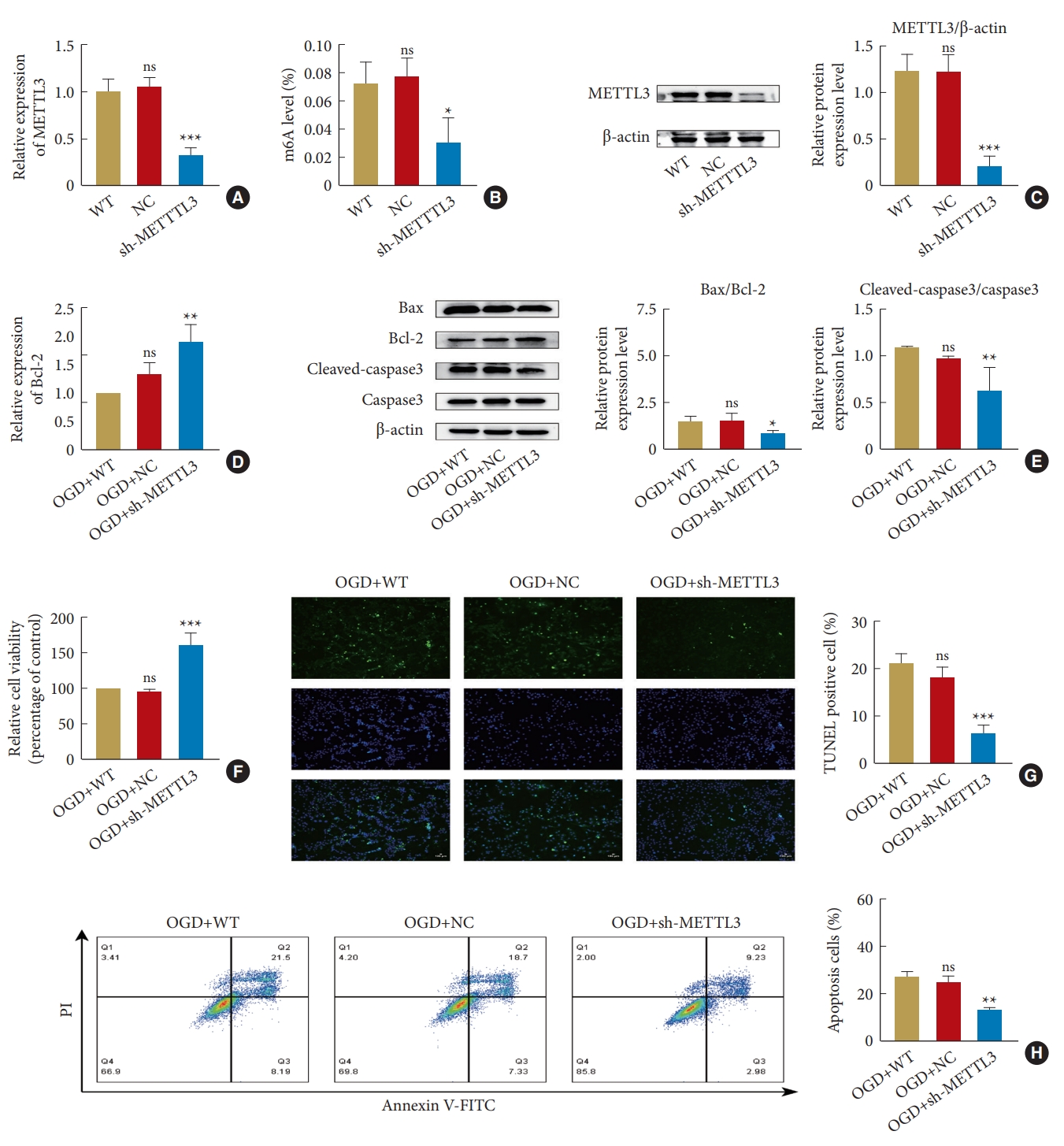
METTL3 Knockdown promotes survival and inhibits apoptosis of PC12 cells after OGD. (A) Reverse transcription quantitative polymerase chain reaction measuring METTL3 transcription after its knockdown. (B) m6A colorimetric assay measuring m6A modification after METTL3 knockdown. (C) Reduced protein expression of METTL3 measured by western blotting after METTL3 knockdown. (D) METTL3 Knockdown increased Bcl-2 transcription under OGD. (E) Western blotting of apoptosis-related protein after METTL3 knockdown. (F) CCK8 assay for measuring cell activity. (G) TUNEL assay for assessing apoptosis. (H) Flow cytometry assay of PC12 cells for apoptosis. WT, wild type; NC, negative control; sh-METTTL3, short hairpin METTTL3; m6A, N6-methyladenosine; OGD, oxygen-glucose deprivation; Bcl-2, B-cell lymphoma 2; mRNA, messenger RNA; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; V-FITC, V-fluorescein isothiocyanate. The significance levels are shown as follows: ns, not significant; versus control group: *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
SCI is associated with increased social costs and a reduced life expectancy [6,54,55]. SCI can be primary and secondary [56]. Recently, accumulating evidence has shown that secondary neuronal injury plays an important but preventable role in the late stages of SCI [57,58]. m6A modification, the most common mRNA modification, and its recognized methylation enzyme, METTL3, play critical roles in various diseases [59]. m6A modification and METTL3 are essential for neuronal physiological functions such as learning and memory [60,61]. METTL3 and m6A levels are upregulated in lung and breast cancer cells [30,31]. However, the role of METTL3 in SCI has not been entirely elucidated.
In the present study, the overall m6A level and METTL3 expression levels gradually increased in PC12 cells after OGD. However, in the rat spinal cord hemisection model, the overall m6A modification levels and METTL3 expression levels reached the maximum on the third day of injury, indicating that SCI is associated with overall m6A modification levels and METTL3 expression levels. Apoptosis induced by SCI exacerbates the extent of injury due to neuronal loss and dysfunction [6,57]. In previous studies, altered m6A modification and increased METTL3 expression have been observed in SCI, and gene transcription levels increased by hypomethylation [62]. In a mouse model of chronic inflammatory pain, the levels of m6A modifications and METTL3 significantly increased in the spinal cord [63]. These studies suggest that METTL3 may play an important role in SCI. Based on changes in METTL3 and m6A in the spinal cord, we hypothesized that METTL3 inhibition can regulate m6A modifications and modulate SCI.
Bcl-2 regulates apoptosis by controlling intracellular signaling pathways [52]. By binding to Bax, Bcl-2 reduces Bax-mediated caspase3 activation and apoptosis; Therefore, Bcl-2 downregulation can enhance apoptosis [64,65]. METTL3 has been reported to catalyze m6A modifications of Bcl-2 in chondrocytes [45]. Bcl-2 has also been indicated as a target gene of METTL3 in lung and breast cancers [30,31]. Similarly, Bcl-2 inhibits neuronal apoptosis [32,66,67]. In our study, bioinformatics revealed the m6A methylation modification sites and common motifs of Bcl-2 mRNA. It has been shown that METTL3 can bind to transcripts in the cytoplasm to form a loop structure, resulting in repeated translation of the protein [46]. Therefore, we suggested that the secondary structure of RNA may have an impact on METTL3 function; therefore, we predicted the secondary structure of Bcl-2 mRNA using the SRAMP database. METTL3 has 580 amino acids and consists of a ZFD domain, which ZFD plays a role in RNA recognition, and a methyltransferase domain [18,47]. Therefore, we analyzed the interaction between Bcl-2 mRNA and METTL3 using the catRAPID database. We found that there are interactions between Bcl-2 mRNA and METTL3 and the binding region of METTL3 is not in the same site as its catalytic region. We used meRIP to confirm the presence of m6A modification on Bcl-2 mRNA, and RIP analysis to show that Bcl-2 mRNA is a downstream target gene of METTL3. There was a significant difference in the enrichment level of Bcl-2 mRNA between the 2 experiments, indicating that Bcl-2 mRNA can be formed by METTL3, but the possible role of other methyltransferases cannot be excluded. Therefore, we hypothesize that the m6A modification of Bcl-2 in SCI is mediated by METTL3 and can affect neuronal apoptosis.
It has been reported in the literature that METTL14, another methylation enzyme, exacerbates the severity of SCI, and m6A modification reduces the expression of genes that inhibit neuronal apoptosis [68,69]. Han et al. [70] revealed a higher m6A methylation of many genes and METTL3 upregulation in Alzheimer disease. Therefore, we further explored the role of METTL3 in neuronal apoptosis in SCI. STM2457, a novel METTL3-specific inhibitor, can bind to the SAM binding site and effectively inhibit METTL3 [71]. We measured the inhibitory effect of STM2457 on METTL3 in PC12 cells. In vitro experiments revealed that the inhibitory effect of STM2457 on METTL3 started from 5 μM, and stabilized after 10 μM, but the cellular activity was fixed between 0 μM and 25 μM. Therefore, we set the effective inhibitory concentration to 10 μM. The results showed that the mRNA and protein levels of Bcl-2 significantly increased in the presence of STM2457, while the expression of other apoptosis-related proteins showed different degrees of decrease. After adding STM2457, the TUNEL-positive PC12 cells were reduced, the neuronal survival rate significantly increased, and the total apoptosis rate was significantly lower in the OGD+STM2457 group than in the OGD group on Annexin V-FITC/PI flow cytometry. These results suggest that STM2457 has an important role in inhibiting PC12 cells apoptosis. Previous studies have also shown that METTL3, m6A methylation, and neuronal apoptosis are closely related [72,73]. Xu et al. [43] found that silencing METTL3 can decrease the m6A methylation level of LNC-D63785, upregulate its expression, and reduce neuronal apoptosis. Therefore, we constructed stable knockdown METTL3 cell lines to investigate the effect of METTL3 on Bcl-2 and PC12 cells apoptosis. After METTL3 knockdown, a 12-hour OGD model was established, and it was found that the transcription and protein expression of Bcl-2 increased in the OGD+sh-METTL3 group, while other apoptosis-related proteins were downregulated. In addition, lower TUNEL-positive cells and apoptosis rates were observed in the OGD+sh-METTL3 group.
In summary, we demonstrated that inhibition of METTL3 activity and expression reduces OGD-induced apoptosis of PC12 cells. Specifically, METTL3 modulates Bcl-2 transcription in an m6A-dependent manner, thereby altering the protein level of Bcl-2, and neuronal apoptosis (Fig. 6). The METTL3/m6A/Bcl-2 axis is a new target for treating SCI. This study did not validate the downstream targets of METTL3 and other possible pathways in vivo, and there are some limitations. Based on the results of this study, we continue our in-depth analysis in our future studies.
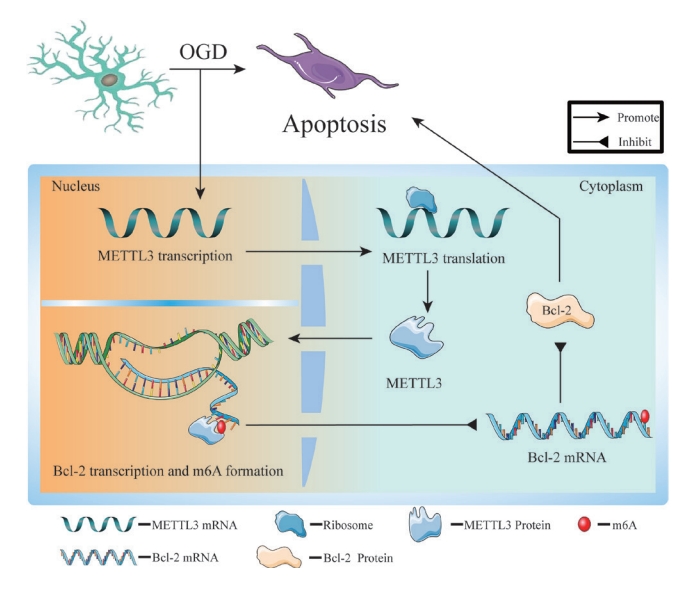
METTL3 forms an m6A modification near the 3ʹ-untranslated region of the Bcl-2 gene, thereby inhibiting Bcl-2 expression and promoting neuronal apoptosis after OGD. Therefore, METTL3 inhibition can upregulate Bcl-2 expression and inhibit neuronal apoptosis. The METTL3/m6A/Bcl-2 axis may provide new insights for targeted therapy in spinal cord injury. m6A, N6-methyladenosine; Bcl-2, B-cell lymphoma 2; mRNA, messenger RNA; OGD, oxygen-glucose deprivation.
Notes
Conflict of Interest
The authors have nothing to disclose.
Funding/Support
We thank the search grant from National Natural Science Foundation of China (Grant number: 82072407), Natural Science Foundation of Fujian Province (Grant number: 2021J01762), Startup Fund for scientific research, Fujian Medical University (Grant number: 2020QH1083), sponsored by Fujian provincial health technology project (Grant number: 2020QN01010187).
Author Contribution
Conceptualization: SG, GC, LZ, ZW; Data curation: SG, TL, LZ, ZC, TS, DC; Formal analysis: SG, TL, ZC, T Shi, DC; Funding acquisition: GC, ZS, WL; Methodology: SG, GC, ZS, ZW, WL; Project administration: ZS, ZC; Visualization: TL, ZS, TS; Writing - original draft: SG; Writing - review & editing: TL, GC.
