Proteomic Comparison of Paraspinal Muscle Imbalance Between Idiopathic Scoliosis and Congenital Scoliosis
Article information
Abstract
Objective
This study aims to compare the proteomic profiles of paraspinal muscle imbalance between idiopathic scoliosis (IS) and congenital scoliosis (CS).
Methods
Bilateral paraspinal muscles of 5 pairs of matched IS and CS patients were collected. Proteome patterns of paraspinal muscles were established. Differentially expressed proteins (DEPs) in paraspinal muscles between the convexity and the concavity were screened out. DEPs shared by both IS and CS and IS-specific DEPs were identified. Bioinformatic analyses of DEPs were performed.
Results
Among 105 DEPs identified in IS, 30 displayed predominant expression on the convexity, whereas other 75 exhibited predominant expression on the concavity. DEPs in IS were mainly enriched in calcium ion binding and DNA binding in gene ontology (GO) term and glycolysis/gluconeogenesis and purine metabolism in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Among 48 DEPs identified in CS, 25 were predominantly expressed on the convexity and 23 on the concavity. DEPs in CS were mainly enriched in receptor activity and immune response in GO term and glycolysis/gluconeogenesis and cellular senescence in KEGG pathway. Comparison of DEPs between IS and CS identified only 8 proteins shared by both types of scoliosis. Among the 97 IS-specific DEPs, 28 were predominantly expressed on the convexity and 69 on the concavity. IS-specific genes were enriched in calcium ion binding and protein glycosylation in GO term and glycolysis/gluconeogenesis and hypertrophic cardiomyopathy in KEGG pathway.
Conclusion
IS and CS exhibit proteomic imbalance in bilateral paraspinal muscles but share few similarities. Paraspinal muscle imbalance in IS might not be the consequence of spinal deformities.
INTRODUCTION
As the most common type of scoliosis, idiopathic scoliosis (IS) affects approximately 1%–3% of the population, which differs between regions and races [1-3]. The course of disease deteriorated progressively in more than 60% of patients [4]. As the most common nonoperative treatment, bracing is effective in preventing curve progression in a certain proportion of IS patients [5,6]. However, brace treatment is mainly used for patients with a Cobb angle of more than 25 degrees. For patients at the early stage of the disease, effective intervention is in lack. To develop early interventions, exploring the cause of IS is essential. Unlike congenital scoliosis (CS), no remarkable vertebra abnormities were observed in IS. Currently, multiple theories have been put forward to interpret the pathogenesis of IS, including the nervous system equilibrium dysfunction theory, hormone theory, genetic theory, etc [7-9]. Nervous system equilibrium dysfunction may bring about the imbalance of paraspinal muscles, the effector of nervous system. The asymmetric distribution and methylation level of estrogen receptors in bilateral paraspinal muscles were observed in IS [10-12]. As for genetic theory, single nucleotide polymorphisms in several genes that were asymmetrically expressed in bilateral paraspinal muscles were identified in IS [13-16]. Based on above theories, paraspinal muscle imbalance is supposed to participate in the pathogenesis of IS.
Various methods were used to decipher paraspinal muscle imbalance in IS. For instance, imageological techniques, such as ultrasound and magnetic resonance imaging (MRI), verified the differences between bilateral paraspinal muscles regarding cross-sectional area and muscle volume [17-20]. Asymmetrical myoelectric activity was observed by electromyography [21]. Histological analyses showed differences between the concavity and the convexity in the size, density and distribution of myofibers [22,23]. Although relevant research has mushroomed, whether paraspinal muscle imbalance is the primary cause or secondary change of scoliosis in IS has yet to be determined. To clarify the causality between paraspinal muscle imbalance and scoliosis, more precise methods are required. Recently, transcriptome sequencing has been used to identify differentially expressed genes (DEGs) in bilateral paraspinal muscles in IS [24,25]. Although several crucial DEGs were identified in IS, the difference of global gene expression profile in bilateral paraspinal muscles between IS and CS remains unclarified. Besides, to our knowledge, research on paraspinal muscle imbalance in IS from the aspect of proteome is in lack.
Herein, we utilized data-independent acquisition (DIA) proteomics to analyze differences in paraspinal muscles between the convex and concave sides in IS. Besides, CS was taken as the control. As is known, differences in paraspinal muscles between the convex and concave sides in CS are supposed to be changes due to genetic factors for skeletal deformities or changes secondary to skeletal deformities. Comparison of paraspinal muscle imbalance between IS and CS may help to eliminate differentially expressed proteins (DEPs) in bilateral paraspinal muscles secondary to scoliosis, and thereby screen out DEPs specific for IS, which may give some enlightenments for the pathogenesis of IS.
MATERIALS AND METHODS
1. Participants
IS patients and CS patients receiving surgical treatment by one senior surgeon (Prof. Shen) from January 2022 to December 2022 in Peking Union Medical College Hospital were enrolled in this study. The inclusion criteria for IS were as follows: (1) Age at surgery ranged from 10 to 18 years old. (2) adolescent idiopathic scoliosis with thoracic curve as the major curve. (3) Primary surgery via posterior spinal instrumentation. The inclusion criteria for CS were: (1) Age at surgery ranged from 10 to 18 years old. (2) Apex of the curve located at T5/6 disc to T11/12 disc. (3) Posterior spinal fusion as the first surgery with no previous nonfusion surgeries or revision surgery. (4) Intraspinal abnormities, including split cord malformation, syringomyelia and tethered spinal cord, were excluded using computed tomography scans and MRI. (5) Without a definite genetic diagnosis. Then IS patients and CS patients were matched according to age (≤ 1 year), sex and the location of apex (≤ 1 segment). Patient selection strategy was described in Fig. 1.
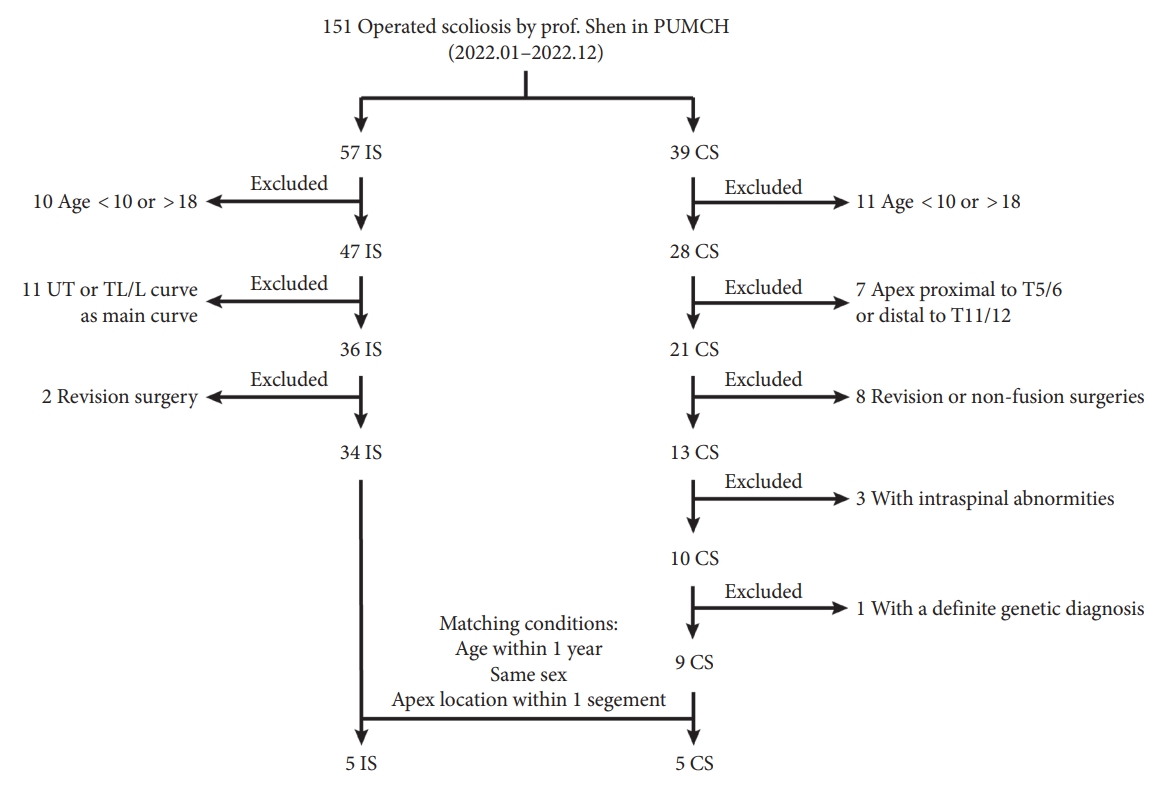
Flowchart showing the recruitment of participants in this study. 5 pairs of IS patients and CS patients were selected from scoliotic patients receiving corrective surgery by Prof. Shen in PUMCH in 2022 and matched according to age, sex and apex location. PUMCH, peking union medical college hospital; IS, idiopathic scoliosis; CS, congenital scoliosis; UT, upper thoracic; TL/L, thoracolumbar/lumbar.
This study was performed according to the Helsinki Declaration and approved by the Institutional Review Board of Peking Union Medical College Hospital (K4136). A written informed consent was obtained from all subjects.
2. Specimens Collection, Processing and Storage
After general anesthesia, patients were lied in prone position. Then a standardized posterior median approach was taken to expose bony structures. Muscle samples were obtained from bilateral multifidus muscles at the apical region of the major curve. To ensure the symmetry of samples collected, deep muscles adjacent to spinous process, which covers vertebral lamina at the apical vertebra were collected. All procedure were conducted by a senior surgeon (Prof. Shen) specialized in spinal deformity.
After collection, muscles were washed with precooled phosphate buffer saline to remove blood cells. Then samples were dissected into suitable size and immersed in AllProtect preservation solution of nucleic acids and proteins in animal tissues (Beyotime Biotechnology, Shanghai, China) at 4°C overnight. Tissues were then transferred to -80°C for storage until use.
3. Protein Extraction, Quality Control, and Digestion
Protein extraction was performed by the filter-aided sample preparation procedure. Briefly, tissues with the size of approximately 3 mm × 3 mm × 3 mm were grinded in liquid nitrogen and lysed with lysis buffer containing sodium dodecyl sulfate (SDS) and dithiothreitol (4% SDS [Sigma-Aldrich, St. Louis, MO, USA], 1 mM DL-Dithiothreitol [Sigma-Aldrich], 100 mM Tris–HCl [Sigma-Aldrich], pH 7.6). After ultrasonication for 5 minutes, the lysates were incubated at 95°C for 15 minutes and with ice-bath for 2 minutes. Then the lysates were centrifuged at 12,000 g for 15 minutes at 4°C to remove insoluble cellular debris. The supernatants were obtained and mixed with sufficient iodoacetamide (Solarbio, Beijing, China). After alkylation at room temperature for 1 hour in darkness, samples were mixed with 4 volumes of precooled acetone (Solarbio) and incubated at -20°C for 120 minutes. Precipitates were collected via centrifugation at 12,000 g for 15 minutes at 4°C, followed by washing with 1-mL precooled acetone (Solarbio). Then the precipitates were completely dissolved in Dissolution Buffer (8 M Urea, Solarbio; 100 mM tetraethylammonium bromide, Solarbio; pH 8.5).
Protein concentration was quantified via the Branford method, according to the manufacturer’s instructions (Beyotime Biotechnology, Shanghai, China). Then samples of 20 μg total protein were used for protein quantification and SDS-poly acrylamide gel electrophoresis (Solarbio).
After quantification, 100 μg total protein of each samples was digested into peptides. Briefly, each protein samples were added to 100-μL dissolution buffer. Samples were then reacted with trypsin and 100 mM tetraethylammonium bromide (Solarbio) at 37°C for 4 hours, followed by digestion with trypsin and calcium chloride (Solarbio) overnight. Then formic acid (FA; Sigma-Aldrich) was added to adjust the pH of digested samples under 3. After centrifugation at 12,000 g for 5 minutes at room temperature, the supernatant was collected and loaded to the C18 desalting column. Then the column was washed with washing buffer (0.1% FA, Sigma-Aldrich; 3% acetonitrile, Sigma-Aldrich) for 3 times. After that, elution buffer (0.1% FA, Sigma-Aldrich; 70% acetonitrile, Sigma-Aldrich) was added and the eluents were collected and lyophilized.
4. Peptide Fractionation
Lyophilized samples were resolved in mobile phase buffer A (2% acetonitrile, Sigma-Aldrich; pH 10). Then samples were centrifugated at 12,000 g for 10 minutes at room temperature. Fractionation were performed using L-3000 HPLC system (Arc Scientific, Syracuse, NY, USA) connected with Waters BEH C18 column (4.6 × 250 mm, 5 μm). Then samples were subjected to the column. Gradient elution was performed with different proportions of mobile phase buffer A and buffer B (98% acetonitrile, Sigma-Aldrich; pH 10). After fractionation, samples were lyophilized, followed by dissolved in 0.1% FA (Sigma-Aldrich).
5. Data-Dependent Acquisition- and DIA-Based LC-MS/MS
The data-dependent acquisition (DDA) and DIA liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analyses were performed using an EASY-nLC 1200 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Samples were added with indexed retention time (iRT) peptides (Biognosys, Schlieren, Switzerland) according to manufacturer’s instructions. Peptides of 1 μg were loaded onto the self-produced precolumn. Gradient elution was then conducted with different proportions of mobile phase buffer A (0.1% FA, Sigma-Aldrich) and buffer B (80% acetonitrile, Sigma-Aldrich; 0.1% FA, Sigma-Aldrich). For DDA mode, parameters were set as follows: full scan range, 350 to 1,500 m/z; MS1 scan resolution, 120,000 at 200 m/z; automatic gain control (AGC) target value, 3 × 106; maximum ion injection time (MIIT), 80 msec. The top 40 precursor ion with the highest abundance were screened out, fragmented using higher energy collisional dissociation, followed by MS2 analysis. Parameters for MS2 were set as follows: scan resolution, 15,000 at 200 m/z; AGC target value: 5 × 104; MIIT, 45 msec; normalized collision energy: 27%.
6. MS Data Analysis
Raw data of DDA were searched against the National Center for Biotechnology Information database (1492129-1492124-Homo_sapiens. fasta; 130184 sequences) using Spectronaut Pulsar X software (Biognosys AG, Schlieren, Switzerland). Parameters for the searches were as follows: mass tolerance for precursor ion, 10 ppm; mass tolerance for product ion, 0.02 Da; maximum of missed cleavage sites, 2; fixed modification, carbamidomethyl; dynamic modification, oxidation of methionine; N-terminal modification, acetylation. Retrieval results were further filtered by Spectronaut Pulsar X software (Biognosys AG) to select identified peptide spectrum matches (PSMs) with the confidence level greater than 99%. Identified PSMs were then subjected to the verification of false discovery rate (FDR) for the removal of peptides and proteins with FDR greater than 1%. Then the DDA spectral library was established.
DIA data was imported into Spectronaut Pulsar X software (Biognosys AG). By search DDA spectral library, qualitative and quantitative analysis of peptides were implemented. Parameters set for DIA analysis were as follows: retention time correction, dynamic iRT; precursor ion Q value cutoff, 0.01.
7. Grouping and Comparison
Samples collected from the convex and concave sides in IS patients were classified as the IST group and ISA group, respectively. To investigate proteomic imbalance of paraspinal muscles in IS, comparison between the IST group and ISA group were conducted. Samples collected from the convex and concave sides in CS patients were classified as the CST group and CSA group, respectively. To investigate proteomic imbalance of paraspinal muscles in CS, comparison between the CST group and CSA group were conducted. Upregulation was defined as a higher expression of DEPs on the convex side than the concave side, whereas downregulation was defined as a lower expression of DEPs on the convex side than the concave side. To determine IS-specific proteomic profile, DEPs in paraspinal muscles between the convex and concave sides in IS were compared with that in CS. After the elimination of DEPs with the same trend in bilateral paraspinal muscles shared by both IS and CS, the remaining DEPs in IS were defined as the IS-specific DEPs.
8. Bioinformatic Analysis
Fold change >1.5 and <0.67 were set to identify DEPs between groups, with a p-value of < 0.05 determined by independent t-test. Volcano plot and hierarchical clustering heatmap with R package (version 3.4.3) were used to analyze DEPs. Gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Interpro (IPR) domain annotation were performed using R ver. 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria) to explore functional enrichment of DEPs. Cytoscape (ver. 3.8.2) were utilized to construct protein-protein interaction (PPI) network and screen out hub proteins.
9. Statistical Analysis
Clinical data were analyzed using IBM SPSS Statistics ver. 22.0 (IBM Co., Armonk, NY, USA). Continuous variables were presented as mean±standard deviation and compared by independent t-test or Mann-Whitney U-test. Categorical variables were presented as percentages (%) and compared by chi-square test. Proteomic data were processed by Spectronaut Pulsar X software (Biognosys AG) with default settings, and the FDR was also set as 1%. Proteomic analysis between groups were compared by independent t-test. A p-value of < 0.05 was considered statistically significant. Hypergeometric test was applied for enrichment analyses of GO terms, KEGG pathways and IPR domains, with a p-value of < 0.05 was considered significant.
RESULTS
1. Patient Characteristics
A total of 5 IS patients and 5 CS patients were matched in this study (Table 1). Average age for IS group and CS group were 13.8±0.8 and 13.0±1.2 years old, respectively, with no statistical difference between groups (p < 0.05). There were 4 females in each group (p < 0.05). No significant differences were observed in major curve-related parameters, including curve length, Cobb angle and the location of apical vertebra (All p < 0.05).
2. Proteomic Analysis of Paraspinal Muscle Imbalance in IS
Comparison of paraspinal muscles on the convex and concave sides identified 105 DEPs in IS (Fig. 2A). Among these DEPs, 30 exhibited higher expression on the convex side than the concave side; while the remaining 75 DEPs displayed lower expression on the convex side than the concave side (Fig. 2B). According to GO annotation of biological process, DEPs in IS were enriched in calcium ion binding, DNA binding, and et al. (Fig. 2C). KEGG analysis revealed the enrichment of DEPs in IS in various pathways, including glycolysis/gluconeogenesis, purine metabolism, hypertrophic cardiomyopathy, et al. (Fig. 2D). IPR domain annotation revealed that DEPs in IS were enriched in Vitamin K epoxide reductase and calsequestrin (Fig. 2E). Subcellular localization of DEPs in IS indicated that 22 proteins localized in cytoplasm, 17 in nucleus, 9 in mitochondrion, 8 in cytoskeleton, 7 in plasma membrane and 5 in centrosome (Fig. 2F). Global analysis of DEPs in IS identified 89 nodes and 405 edges involved in PPI network (Fig. 2G). The top 10 hub proteins were myosin light chain 11 (MYL11), troponin C2, fast skeletal type (TNNC2), tropomyosin alpha-1 chain isoform Tpm1.2st (TPM1.2st), troponin I2, fast skeletal type (TNNI2), actinin alpha 3 (ACTN3), ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 (ATP2A1), myosin light chain 1 (MYL1), nebulin (NEB), myozenin 1 (MYOZ1), and myosin-binding protein C2 (MYBPC2) (Fig. 2H).

Comparison of proteomic profiles between the IST group and ISA group. (A) Hierachical clustering heatmap based on 105 DEPs in IS. (B) Volcano plot shows that 75 DEPs were highly expressed in the ISA group and 30 were highly expressed in the IST group. (C) Gene ontology (GO) analysis indicates functional enrichment of DEPs in IS in biological processes, such as protein glycosylation, and molecular functions including calcium ion binding and DNA binding. (D) Kyoto Encyclopedia of Genes and Genomes analysis reveals the enrichment of DEPs in IS in pathways, including glycolysis/gluconeogenesis, purine metabolism, etc. (E) IPR analysis indicates the enrichment of DEPs in IS in Vitamin K epoxide reductase, calsequestrin, et al. (F) Subcellular distribution of DEPs in IS. (G) Protein-protein interaction network of DEPs in IS identified 89 nodes and 405 edges. (H) Top 10 hub proteins in protein-protein interaction network of IS. IST, paraspinal muscles on the convex side in idiopathic scoliosis; ISA, paraspinal muscles on the concave side in idiopathic scoliosis; IS, idiopathic scoliosis; DEP, differentially expressed protein; BP, biological process; MF, molecular function; PWWP, Pro-Trp-Trp-Pro motif.
3. Proteomic Analysis of Paraspinal Muscle Imbalance in CS
Forty-eight DEPs in bilateral paraspinal muscles were detected in CS patients (Fig. 3A). Higher expression of 25 DEPs on the convex side than the concave side were observed. The remaining 23 DEPs exhibited lower expression on the convex side than the concave side (Fig. 3B). GO analysis revealed that DEPs in CS were enriched in receptor activity in the category of molecular function and immune response in the category of biological process (Fig. 3C). Analysis of KEGG pathway suggested the enrichment of DEPs in CS in glycolysis/gluconeogenesis, cellular senescence, Kaposi’s sarcoma-associated herpesvirus infection and biotin metabolism (Fig. 3D). IPR domain annotation showed that the enrichment of DEPs in CS in various domains including methyltransferase, NNMT/PNMT/TEMT, phosphoglycerate kinase, frataxin/CyaY, etc. (Fig 3E). Subcellular localization of DEPs in CS revealed the distribution of 9 proteins in cytoplasm, 6 in nucleus, 4 in plasma membrane, 4 in cytoskeleton, 3 in mitochondrion and 3 in extracellular space (Fig. 3F). PPI network of DEPs in CS displayed 43 nodes and 69 edges (Fig. 3G). The top 10 hub proteins were glyceraldehyde-3-phosphate dehydrogenase, titin, myosin heavy chain 4, parvalbumin, protein phosphatase 3 catalytic subunit alpha, PDZ (PSD-95, Dlg, and ZO-1/2) and LIM (Lin11, Isl- 1, and Mec-3) domain 7, succinate-CoA ligase guanosine diphosphate-forming subunit beta, cullin 4A, cyclin dependent kinase 6 and phosphoglycerate kinase 1 (PGK1) (Fig. 3H).
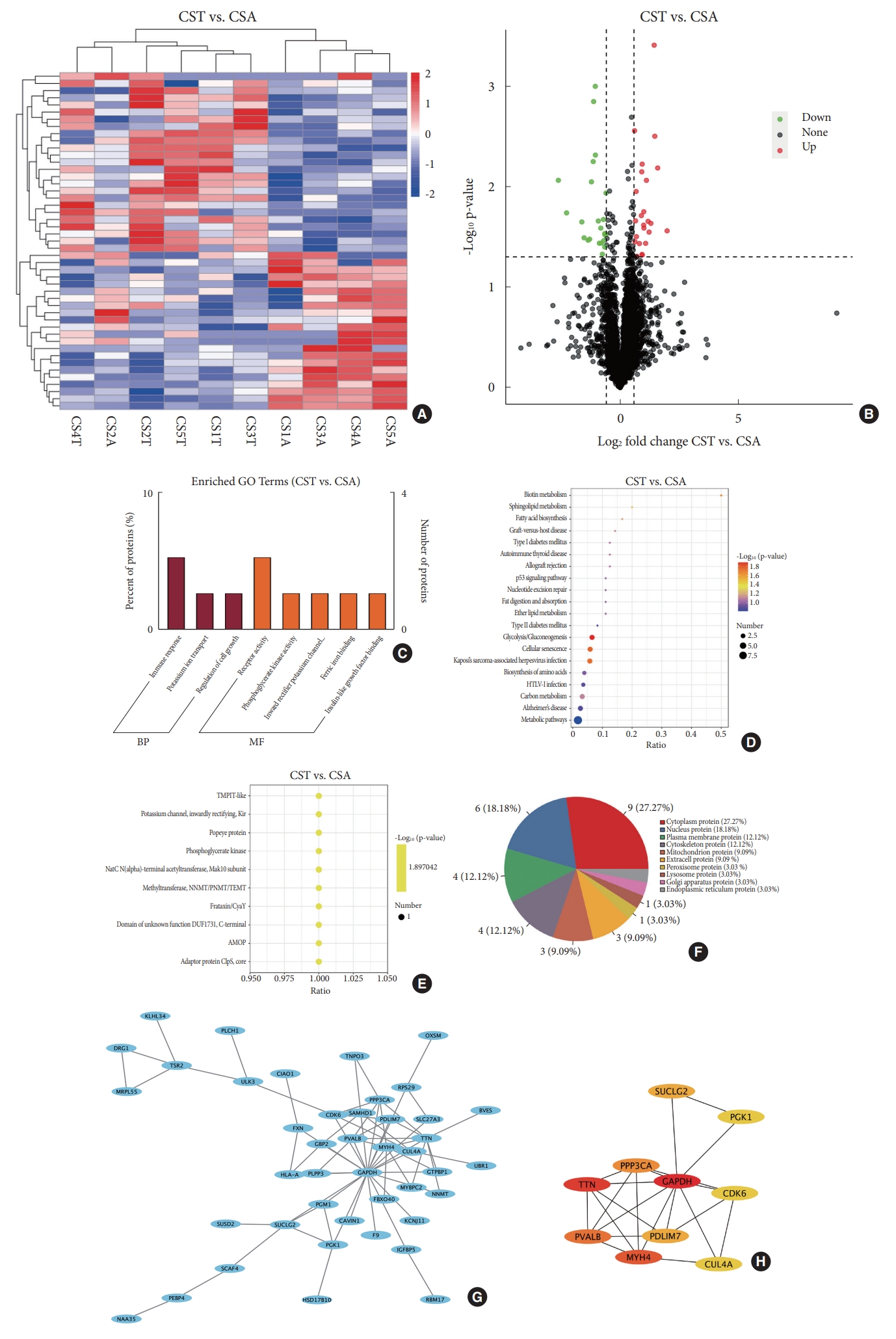
Comparison of proteomic profiles between the CST group and CSA group. (A) Hierachical clustering heatmap based on 48 DEPs in CS. (B) Volcano plot shows that 25 DEPs were highly expressed in the CSA group and 23 were highly expressed in the CST group. (C) Gene ontology (GO) term analysis indicates functional enrichment of DEPs in CS in immune response and receptor activity. (D) Kyoto Encyclopedia of Genes and Genomes pathway analysis reveals the enrichment of DEPs in CS in glycolysis/gluconeogenesis, cellular senescence, etc. (E) IPR domain analysis exhibits the enrichment of DEPs in CS in methyltransferase, NNMT/PNMT/TEMT, phosphoglycerate kinase, et al. (F) Subcellular distribution of DEPs in CS. (G) Protein-protein interaction network of DEPs in CS identified 43 nodes and 69 edges. (H) Top 10 hub proteins in protein-protein interaction network of CS. CST, paraspinal muscles on the convex side in congenital scoliosis; CSA, paraspinal muscles on the concave side in congenital scoliosis; CS, congenital scoliosis; DEP, differentially expressed protein; BP, biological process; MF, molecular function; TMPIT, transmembrane protein induced by tumor necrosis factor alpha; NNMT/PNMT/TEMT, nicotinamide N-methyltransferase/phenylethanolamine Nmethyltransferase/thioether S-methyltransferase; DUF1731, domain of unknown function 1731; AMOP, adhesion-associated domain in MUC4 and other proteins.
4. Identification of DEPs in Bilateral Paraspinal Muscles Shared by Both IS and CS
Comparison of DEPs in bilateral paraspinal muscles in IS and that in CS was performed. Only 8 DEPs with same trend in both IS and CS were identified (Table 2). Among DEPs shared by both IS and CS, 6 exhibited lower expression in paraspinal muscles on the convex side than that on the concave side; while the expression of the remaining 2 DEPs were higher on the convex side than that on the concave side in IS (Fig. 4A, B). GO annotation demonstrated that DEPs shared by both groups were enriched in protein binding, carbohydrate metabolic process and organic substance catabolic process (Fig. 4C). KEGG pathway analysis revealed DEPs shared by both groups were enriched in glycolysis/gluconeogenesis, nucleotide excision repair, galactose metabolism, et al. (Fig. 4D). IPR domain annotation showed the enrichment of DEPs shared by both groups in phosphoglycerate kinase, PDZ domain, alpha-D-phosphohexomutase, C-terminal, et al. (Fig. 4E). Among DEPs shared by both groups, 3 were localized in cytoplasm, 3 in nucleus and 2 in cytoskeleton (Fig. 4F). A tight correlation between PGK1 and phosphoglucomutase-1 isoform 1 was observed in PPI network, which corresponds with the enrichment of DEPs in carbohydrate metabolic process in GO annotation and glycolysis/gluconeogenesis pathway in KEGG analysis.
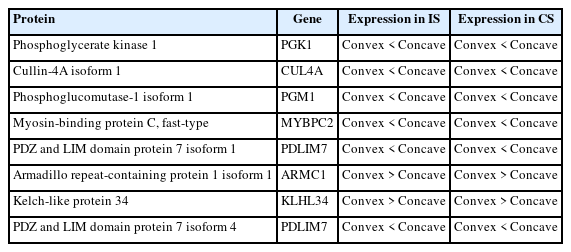
Differentially expressed proteins with the same trend in bilateral paraspinal muscles of both IS and CS
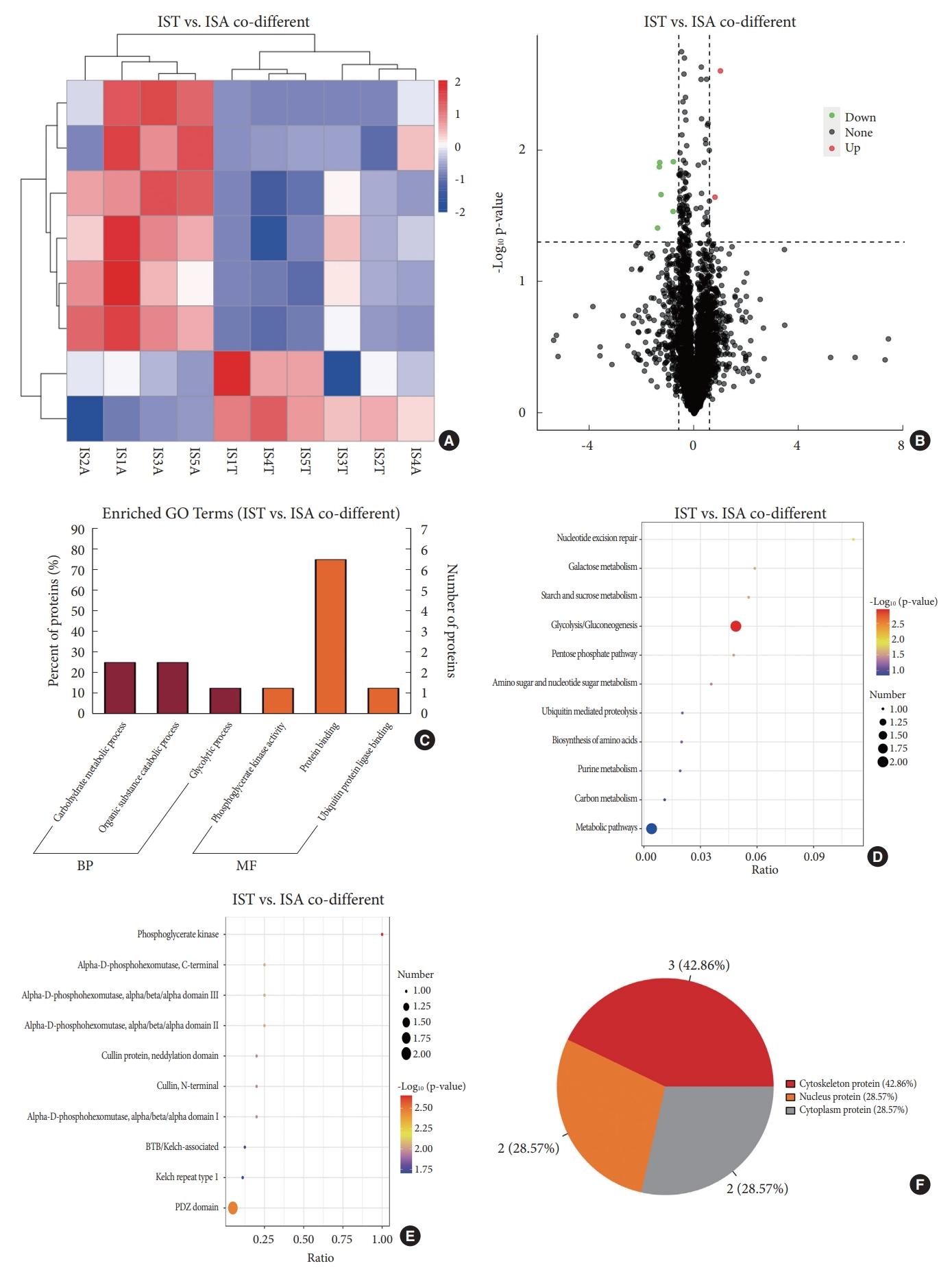
Bioinformatic analysis of DEPs shared by IS and CS. (A) Hierachical clustering heatmap based on 8 DEPs shared by IS and CS. (B) Volcano plot shows that 2 DEPs shared by IS and CS were highly expressed in the IST group and 6 were highly expressed in the ISA group. (C) Gene ontology (GO) analysis indicates functional enrichment of DEPs shared by IS and CS in protein binding, carbohydrate metabolic process, etc. (D) Kyoto Encyclopedia of Genes and Genomes analysis reveals the pathway enrichment of DEPs shared by IS and CS in glycolysis/gluconeogenesis, nucleotide excision repair, etc. (E) Interpro analysis exhibits domain enrichment of DEPs shared by IS and CS in phosphoglycerate kinase, PDZ domain, etc. (F) Subcellular distribution of DEPs shared by IS and CS. IST, paraspinal muscles on the convex side in idiopathic scoliosis; ISA, paraspinal muscles on the concave side in idiopathic scoliosis; IST. vs. ISA shared, differentially expressed proteins in paraspinal muscles between the convex and concave sides in idiopathic scoliosis shared by congenital scoliosis; DEP, differentially expressed protein; IS, idiopathic scoliosis; CS, congenital scoliosis; BP, biological process; MF, molecular function; BTB, broad-complex, tramtrack and bric a brac; PDZ, PSD-95, Dlg, and ZO-1/2.
5. Identification of DEPs in Bilateral Paraspinal Muscles Specific for IS
Comparison of DEPs in bilateral paraspinal muscles in IS and that in CS identified 97 DEPs specific for IS, which account for 91.5% of all DEPs in paraspinal muscles between the convex side and concave side in IS (Fig. 5A). Among those IS-specific DEPs, 69 exhibited lower expression on the convexity than the concavity; while the expression of the remaining 28 DEPs were higher on the convexity than that on the concavity (Fig. 5B). GO analysis indicated that these IS-specific DEPs were enriched in calcium ion binding, protein glycosylation, DNA binding, et al. (Fig. 5C). KEGG pathway analysis showed the enrichment of IS-specific DEPs in multiple pathways, including glycolysis/gluconeogenesis, hypertrophic cardiomyopathy, purine metabolism, et al. (Fig. 5D). IPR annotation exhibited the enrichment of IS-specific DEPs in functions including Vitamin K epoxide reductase, calsequestrin, etc. (Fig. 5E). Subcellular localization of IS-specific DEPs showed that 20 proteins distributed in cytoplasm, 15 proteins in nucleus, 9 in mitochondrion, 7 in plasma membrane, 5 in the cytoskeleton, and 5 in the centrosome (Fig. 5F). Comprehensive analysis of IS-specific DEPs identified a total of 82 nodes and 350 edges involved in PPI network (Fig. 5G). The top 10 hub proteins were MYL11, TNNC2, TPM1.2st, TNNI2, ATP2A1, ACTN3, MYL1, NEB, MYOZ1, and tropomyosin alpha-1 chain isoform Tpm1.5cy (TPM1.5cy) (Fig. 5H).
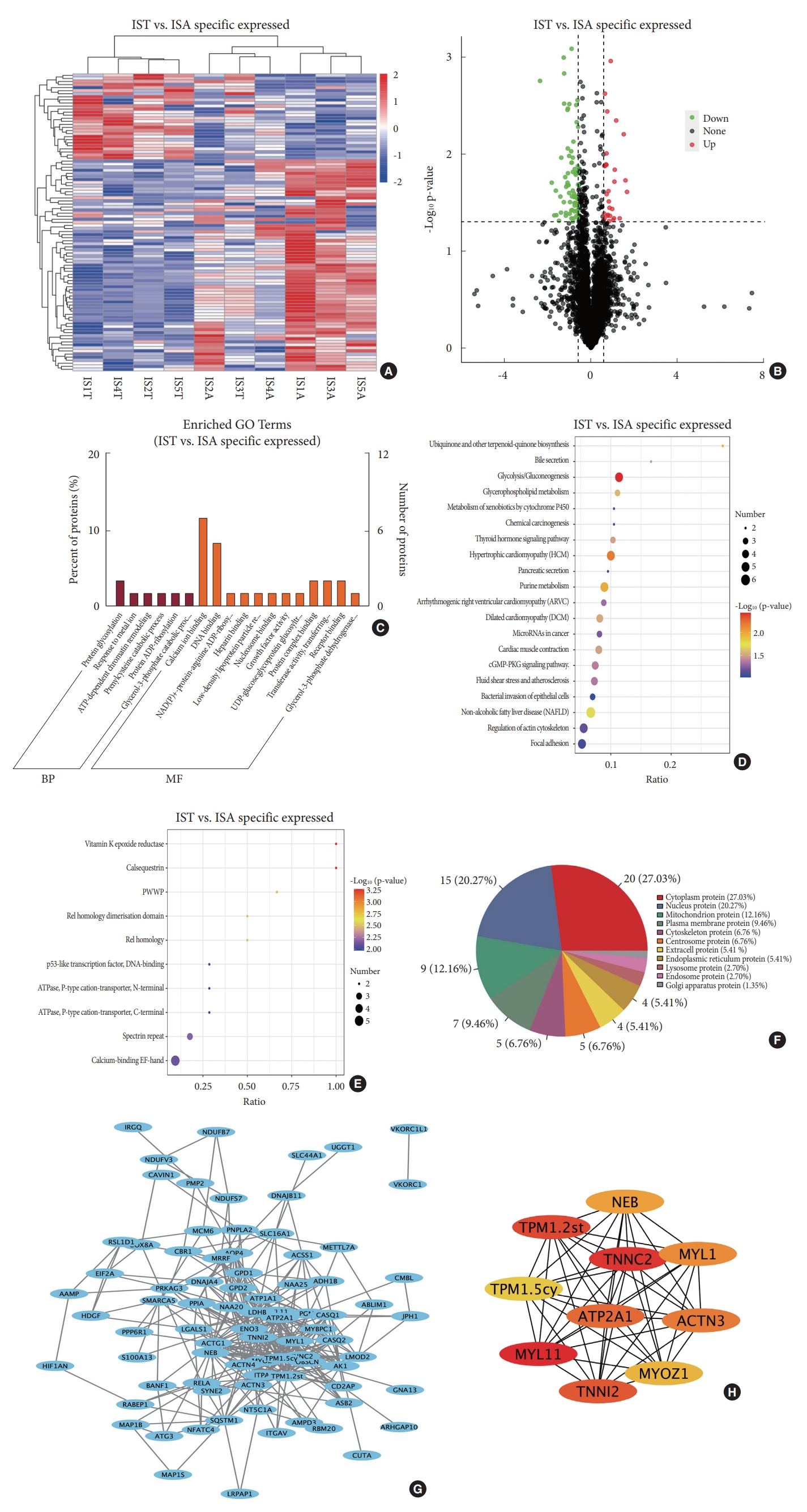
Bioinformatic analysis of IS-specific DEPs. (A) Hierachical clustering heatmap based on 97 IS-specific DEPs. (B) Volcano plot shows 69 DEPs were highly expressed in the ISA group and 28 were highly expressed in the IST group. (C) Gene ontology (GO) analysis indicates functional enrichment of IS-specific DEPs in biological processes, such as protein glycosylation, and molecular functions, such as calcium ion binding, protein glycosylation, etc. (D) Kyoto Encyclopedia of Genes and Genomes analysis reveals the pathway enrichment of IS-specific DEPs in glycolysis/gluconeogenesis, hypertrophic cardiomyopathy, purine metabolism, etc. (E) Interpro analysis exhibits domain enrichment of IS-specific DEPs in Vitamin K epoxide reductase, calsequestrin, et al. (F) Subcellular distribution of IS-specific DEPs. (G) Protein-protein interaction network of IS-specific DEPs identified 82 nudes and 350 edges. (H) Top 10 hub proteins in protein-protein interaction network of IS-specific DEPs. IST. vs. ISA specific, differentially expressed proteins in paraspinal muscles between the convex and concave sides specific in idiopathic scoliosis; DEP, differentially expressed protein; IS, idiopathic scoliosis; BP, biological process; MF, molecular function; BTB, broad-complex, tramtrack and bric a brac; PWWP, Pro-Trp-Trp-Pro motif.
DISCUSSION
This study first described paraspinal muscle imbalance in IS and CS from the aspect of proteome. Comparison of DEPs in paraspinal muscles between the concave and convex sides revealed low similarity in DEPs between IS and CS. Therefore, it is inferred that paraspinal muscle imbalance in gene expression in IS seems not to be changes secondary to the deformity.
Researches about paraspinal muscle imbalance in IS have emerged since decades ago. General observation during surgery indicates the imbalance of muscle volume and fatty infiltration in bilateral paraspinal muscles. To verify this discovery, various imaging methods were undertaken. For instance, ultrasound examination indicated a greater muscle thickness on the concavity [18]. Detection of paraspinal muscles in IS with MRI showed a greater cross-sectional area and muscle volume on the concavity [19,20]. MRI also demonstrated more severity of fatty infiltration on the concavity than that on the convexity [26]. Except for asymmetrical distribution between the concave side and convex side, fatty infiltration in paraspinal muscles of IS also has other typical properties. The severity of fatty infiltration on the concavity of the curve increases gradually from the end vertebra towards the apex [27]. A significant effect of the medial-to-lateral distribution was also observed, with more severe fatty infiltration in the medial (multifidus) than the lateral (erector spinae). Overall, multifidus at the apex on the concavity was speculated as the most severely affected area. Therefore, multifidus muscles at the apical region were selected as the main focus in this study. In addition to conventional imaging modalities, novel technologies were also used to evaluate paraspinal muscle imbalance in IS. X-ray fluorescence demonstrated the asymmetric distribution of metal ions in bilateral paraspinal muscles of IS patients, with higher concentrations of calcium, zinc and copper, on the concavity than the convexity [28]. Detection of back muscles with infrared thermography in IS revealed higher infrared emissivity and temperature on the concavity than the convexity [29]. However, whether CS patients share similar imbalance in above parameters has not been elucidated. Different from imaging methods, histological examinations analyzed paraspinal muscle imbalance in IS from microcosmic aspect. Muscles on the convexity possess a greater cross-sectional area and myonuclei density than that on the concavity [22]. Asymmetrical distribution of myofiber types in bilateral paraspinal muscles was also observed, with a higher proportion of type I fiber and lower proportion of type II fiber on the convexity [23,25,30]. Besides, higher levels of fatty involution and fibrosis were observed on the concavity [31]. Yet histological comparison of paraspinal muscle imbalance between IS and CS is lacking. Electromyogram was widely applied to evaluate myoelectric activity of paraspinal muscles. Despite of the existence of some contrary reports, most studies reported that electromyogram parameters, such as motor unit action potential and root mean square amplitude, on the convexity were higher than that on the concavity in IS [32-34]. Nevertheless, researches on myoelectric activities of paraspinal muscles in CS are rare.
The development of molecular biotechnology provides a possibility to resolve paraspinal muscle imbalance in IS from the aspect of gene expression. Genetic association studies identified exceeding 20 susceptible genes in IS. Some of those genes exhibit differential expression in bilateral paraspinal muscles in IS. For instance, Xu et al. [13] demonstrated a higher expression of LBX1 on the convexity than that on the concavity. However, no differential expression of LBX1 in bilateral paraspinal muscles of CS patients were observed. By targeting MyoD to regulate myogenesis, LBX1 is implicated in the etiology of IS. Similar to LBX1, expressions of other genes in Wnt pathway, such as CTNNB1 and PAX3, show similar difference in IS but no significant difference in CS [35,36]. In addition to genes in Wnt pathway, the expression of SOCS3 in IS and CS was also reported. In IS patients, paraspinal muscles on the convexity exhibited a higher expression of SOCS3 than the concavity, whereas no difference in SOCS3 expression in bilateral paraspinal muscles was observed in CS patients [14]. Unlike above genes, the expression of GPR126 was higher in paraspinal muscles on the concavity than that on the convexity in IS [15]. Howbeit, GPR126 expression in paraspinal muscles did not differ between the concave side and the convex side in CS.
Apart from genomic technologies, transcriptome sequencing was also applied for the detection of paraspinal muscle imbalance in IS. Jiang and colleagues identified 40 DEGs between the concave and convex sides of paraspinal muscles in IS [24]. Of the 40 DEGs, ADIPOQ exhibited a higher expression on the concavity than the convexity in IS, whereas the expression of H19 was higher on the convexity than the concavity. However, neither ADIPOQ or H19 displayed differential expression between the concaved and convexed sided paraspinal muscles in CS. Moreover, differential expression of ADIPOQ and H19 in bilateral paraspinal muscles is correlated with curve severity and the age at initiation. Luo et al. [25] also performed transcriptome analyses and identify 58 DEGs of bilateral paraspinal muscles in IS. They demonstrated that TENT5A expression was higher on the convex side than that on the concave side. Although the pivotal role of TENT5A in myogenesis and muscle fiber maturation, whether CS shares similar differential expression of TENT5A in paraspinal muscles between the concave and convex sides with IS still remains unclear.
As is known, proteins are the main executors of cellular functions. Herein, we applied DIA proteomic sequencing to decipher paraspinal muscle imbalance in IS. GO analysis indicated the enrichment of DEPs in this study in calcium ion binding and DNA binding, whereas DEGs identified in the research of Luo and colleagues mainly enriched in extracellular space, glucose metabolic process, and glucose homeostasis [25]. KEGG analysis demonstrated that DEPs in this study enriched in glycolysis/gluconeogenesis, in line with the report of Jiang et al. [24] IPR domain analysis indicated the enrichment of DEPs in IS in calsequestrin. Calsequestrins participate in muscle contraction through the regulation of calcium storage in the sarcoplasmic reticulum. Calsequestrin 1 is mainly expressed in type II myofibers, whereas calsequestrin 2 is mainly expressed in type I myofibers and cardiomyocytes. In this study, a higher expression of calsequestrin 1 and a lower expression of calsequestrin 2 were observed on the concavity, which may to some degree reflect the asymmetric side distribution of different types of myofibers in IS. All the top 10 hub proteins in the PPI network of DEPs in IS are involved in muscle functions, including muscle contraction and myofibrillogenesis.
It should be pointed out that proteomic analysis of paraspinal muscle imbalance in IS patients only failed to distinguish potential primary DEPs from DEPs secondary to the deformity. To screen out DEPs secondary to scoliosis, bilateral paraspinal muscles in CS were also subjected to proteomic sequencing. It is widely acknowledged that paraspinal muscle imbalance in CS is secondary to the primary vertebral malformations, though genetic causes for skeletal deformities may also bring about muscle lesions in CS. Enrichment of DEPs in glycolysis/gluconeogenesis pathway was observed in CS, similar to that in IS. Besides, DEPs are also enriched in other pathways, including cellular senescence, Kaposi’s sarcoma-associated herpesvirus infection and biotin metabolism. GO analysis showed that DEPs in CS were enriched in immune response and receptor activity. Among the top 10 hub proteins in the PPI network of DEPs in CS, titin, myosin heavy chain 4 and parvalbumin are involved in muscle contraction, whereas PDZ and LIM domain 7 participates in actin cytoskeleton organization.
Obviously, functional enrichments of DEPs in IS are quite different from that in CS. In fact, there is low similarity in DEPs between IS and CS, with only 7.6% of DEPs in IS shared by CS. Except for 8 DEPs shared by both IS and CS, other DEPs in IS were deemed to be specific. These IS-specific DEPs are enriched in calcium ion binding and DNA binding in GO annotation and glycolysis/gluconeogenesis in KEGG pathway. Considering that almost all hub proteins in the PPI network of DEPs in IS are enriched in muscle contraction, it is inferred that the process of muscle contraction, which depends on the release of calcium from sarcoplasmic reticulum and energy supply, may play an important role in paraspinal muscle imbalance in IS.
This study provides comprehensive proteomic landscapes of paraspinal muscles and identified proteomic characteristics of paraspinal muscle imbalance in both IS and CS. Further comparison of DEPs between IS and CS helps to exclude changes in bilateral paraspinal muscles secondary to spinal deformities, which set great obstacles for clarification of the causal relationship between paraspinal muscle imbalance and IS. The identification of DEPs specific for IS provides the basis for future study exploring pivotal genes for paraspinal muscle imbalance in IS and the role of paraspinal muscle imbalance in the pathogenesis of IS. Through restoring the expressions of pivotal genes, targeted therapy may be able to correct paraspinal muscle imbalance, thereby preventing the progression of IS. However, this study still has several limitations. First, sample sizes in this study are relatively small, though patients in IS group and CS group were well-matched regarding age, sex and apex location. Second, this study only identified proteomic differences in paraspinal muscle imbalance between IS and CS, and thus offer some indirect basis for the speculation that paraspinal muscle imbalance might be the cause of IS rather than the consequence. Third, this study only provided a general landscape of differential proteome specific for paraspinal muscle imbalance in IS, further research is required to identified key proteins responsible for paraspinal muscle imbalance in IS.
CONCLUSION
This study demonstrates that differences in proteomic profile in paraspinal muscles between the convex and concave sides are common in both IS and CS. However, IS shares low similarity in DEPs in bilateral paraspinal muscles with CS, suggesting that spinal deformities may not be the principal cause of paraspinal muscle imbalance in IS.
Notes
Conflict of Interest
The authors have nothing to disclose.
Funding/Support
This study was supported by the National Natural Science Foundation of China (grant numbers: 82230083, 81772424), and the Beijing Municipal Science and Technology Project (Z20111000540000).
Author Contribution
Conceptualization: JS; Formal Analysis: ZW, XH; Investigation: ZW; Methodology: ZW, HT, JL, ZL; Project Administration: JS; Writing - Original Draft: ZW; Writing - Review & Editing: XH, HT, JL, ZL, JS.
Acknowledgements
We thank all participants and their guardian for their contributions to this work.

