Predictive Analytics in Spine Oncology Research: First Steps, Limitations, and Future Directions
Article information
Abstract
The potential of big data analytics to improve the quality of care for patients with spine tumors is significant. At this moment, the application of big data analytics to oncology and spine surgery is at a nascent stage. As such, efforts are underway to advance data-driven oncologic care, improve patient outcomes, and guide clinical decision making. This is both relevant and critical in the practice of spine oncology as clinical decision making is often made in isolation looking at select variables deemed relevant by the physician. With rapidly evolving therapeutics in surgery, radiation, interventional radiology, and oncology, there is a need to better develop decision-making algorithms utilizing the vast data available for each patient. The challenges and limitations inherent to big data analyses are presented with an eye towards future directions.
INTRODUCTION
Over the last several decades, there have been major advances in the treatment of primary and metastatic spinal tumors. With continued innovations in surgical techniques and technologies, surgeons are now better equipped to treat the wide spectrum of spinal tumors, whether for palliative reasons or for radical resection and reconstruction. Innovations in implant technology as well as computer-based navigation systems have helped increase the accuracy and durability of surgical reconstructions for spine tumors. Along with these advances, the evolution of cancer-specific therapies in medical and radiation oncology now provide patients with more treatment options, thereby increasing survival [1,2].
As the field of medical oncology advances, the interventions with which patients are being treated are also evolving. Historically, the primary interventions for spinal tumors were primarily radiation and surgery. With innovations in image guidance technology and percutaneous approaches such as cryoablation, cement augmentation, and laser interstitial therapy, the options for treatment are now many and cross multiple disciplines. The incorporation of advances in radiosurgery along with targeted immunotherapies matched to the genomic profiles of the tumors allows for the delivery of precise and personalized care to spine oncology patients [3]. Creating algorithms for clinical decision making is therefore challenging as the interventions are quite broad in scope.
Regardless of tumor type, the goals of treatment are similar in many respects: palliation of pain, preservation or improvement of function, local tumor control, maximizing survival, and improving quality of life. For every patient with a spine tumor, the various treatments are weighed in some fashion to achieve these goals, yet given the myriad variables involved, the decision-making process is confounded by lack of precise predictive data. As such, efforts to study clinical outcomes and associated factors are accelerating at an unprecedented pace. Great advances have been made in other areas of spine surgery, such as spinal deformity surgery, to develop predictive algorithms that can help surgeons and patients alike in the decision-making process [4,5]. For example, predictive algorithms that can help calculate potential complications, readmission, and the invasiveness of surgery may help patients decide on whether to proceed with surgery. Such information would provide a true risk assessment. Similarly, analytics can be performed for surgeon specific questions such as what type of surgery to recommend, how long the fusion construct should be, single stage or multistage surgery. Utilizing large data sets, patterns can be identified, and patientspecific narratives can be created.
In the cancer realm, this has tremendous significance as accurate, precise, and reliable predictive algorithms would help patient and surgeon alike quantify not only the potential benefit of intervention, but the outcomes and impact on quality of life. A patient with metastatic epidural spinal cord compression who has not been able to move his legs or walk for days will want to know if surgery will make a difference. Surgeons currently do not have enough data to provide such counseling to the patient. In fact, current practice often entails mainly reviewing magnetic resonance imaging (MRI) or computed tomography (CT) imaging, assessing the degree of the patient’s weakness or functional impairment, and quantifying the duration of such symptoms. Predicting the potential benefit is difficult and is not necessarily data-driven. Similarly, predicting potential complications after surgery is nearly impossible for a surgeon to do with the millions of data points in the patient’s health record. The patient may be quoted historical data, but how else is the patient supposed to decide about whether to proceed with surgery when they are already faced with a terminal prognosis? This is just one example of how predictive analytics could have an immediate impact at the bedside.
LIMITATION OF THE LITERATURE: SELECTION BIAS
Although the amount of data generated from clinical practice in oncology is immeasurable, published studies are consistently demonstrating gaps in quality of care, high variation in patient outcomes, and costs of care [6]. There are many variations in treatment among surgeons and hospitals. Most of what is referenced in spine oncology is limited by bias, specifically selection bias. Despite this, predictive models keep coming forth based on what has been retrospectively reported and analyzed [7-14].
Why is this important in spine oncology? The selection bias in the literature is a major limitation to the current field. To highlight this issue, consider the example of metastatic epidural spinal cord compression. In many surgical series, outcomes related to surgery for spinal cord compression are taken from an analysis of who had the surgery. Most published reports are retrospective analyses over years at a single institution, often patients treated by the senior physician or surgeon. The problem here is that there are 2 layers of selection bias that have not been accounted for. The first level of bias is how the patient was selected or referred to the surgeon. Someone had to decide whether the patient was suitable for referral. This could be the internist, radiation oncologist, emergency physician, nurse, or medical oncologist. Someone had to make the call and determine that the patient was a surgical candidate. But how did that person decide? And what criteria were used to determine if the patient was a surgical candidate? And what about the other patients that were not referred? How many were referred to radiation? Or even hospice?
The next level of bias is surgeon bias. For every patient reported, there are likely others that were not selected for surgery by the surgeon for whatever reason. Too sick, too old? How a surgeon selects a patient for surgery is certainly not uniform and the driver for decision making is not clear or known, thus highlighting the importance of higher-level predictive analytics using prospective data.
BIG DATA: WHAT IS IT?
The concept of big data is characterized by the 4V’s: volume, velocity, variety, and veracity [15]. For big data analysis to draw meaningful conclusions, researchers should have access to a large volume of data from different sources that will constitute the most possible heterogenous data available and provide an accurate depiction of the real world.
Development of algorithms focuses mainly on the collection of data, and more data. The concept of volume represents the size of a dataset available to run a prediction analysis and extends the classical notion of data available from case reports, small cohort studies or clinical trials, to other diverse sources of available data from electronic health records, imaging, genomics, biosensor data, environmental interactions, wearable technologies, and even social media. The quantification of these interactions and inputs that are being generated in real time will help capture and aggregate big volumes of data to drive insight about health care and clinical outcomes.
In parallel, the concept of velocity refers to the increasing speed with which new data is generated to become available and to be incorporated in available models in order to optimize outcomes. Therefore, it provides prospective information and allows trend prediction if they are computed in a fast and ongoing fashion.
When we think about big data, we think mainly about quantity and large volume, presumably because bigger is often better for data and analytics, but when it comes to clinical data this is not always the case. The real success of big data analytics is not simply driven by processing and managing big data volumes but integrating more sources of data than ever before. This variety challenge aims to enrich the quality of the data collected and reduces bias by guaranteeing more heterogeneity and better representation of the real data.
In addition to volume and heterogeneity of data sources, veracity is a critical feature of data in healthcare. The veracity of data is essential to the level of certainty that what is collected is accurate and therefore the conclusions that are drawn from this data are true and highly accurate. The idea of “garbage in, garbage out” is hallmark because machine learning outputs are based on the quality and accuracy of the data that was provided.
When applied to spine oncology care, big data analytics confer combining and analyzing large datasets to identify associations and make predictions of clinical outcomes. Big data analytics can also be applied to spine imaging, novel therapeutics technologies, drug discovery and delivery, to produce a comprehensive data-driven approach for the treatment and management of spine tumors.
The success of predictive algorithms largely depends on feature selection and data representation. Consider the example of predicting the rate of readmission following surgery for spinal metastatic disease. This is a difficult problem where machine learning has shown some promise using statistical methods [16]. To start, numerous variables including demographics, laboratory variables, surgical parameters, physical performance and comorbidities, imaging studies, intensive care unit stay, functional recovery, and others are collected. With all these data points, it is at first, difficult to decide which variables should be included in the predictive model. Techniques such as univariate analysis, forward or backward step-wise regression models are commonly used to decide upon the variables that should be included. Unfortunately, these variable selection methods lead to algorithms that fail to validate in other datasets and are not generalizable. More advanced machine learning methods such as unsupervised learning (discussed below) can capture interactions between variables to build a more generalizable model. In the following sections we will discuss the available prediction models in spine oncology, focus on the limitations of these models, and guide future directions for big data analytics and prediction in spine oncology.
INTRODUCTION TO ARTIFICIAL INTELLIGENCE
Artificial intelligence (AI) is a branch of computer science that aims to produce tools that can take the capabilities of human intelligence and even outperform human processing tasks when analyzing big data. AI engineering refers to the general ability to design software/machines that independently replicate the intellectual processes of human cognition in deciding on an action in response to its perceived environment to achieve a predetermined goal. In spine surgery, AI is already being used in radiologic diagnosis, surgical planning, and outcome prediction. A straight-forward example of AI application in spine oncology is the development of an automated-segmentation system to identify the vertebrae, neural and vascular elements from CT and MRI to automatically determine the degree of spinal cord compression or the presence of a vertebral fracture [17,18]. An extended application of AI in this area would also be to automate the detection of spinal instability, which could potentially reduce the variability in interpretation among physicians and potentially prompt either surgical referral or closer radiographic follow-up.
Machine learning is a mathematical and computer science statistical tool that focuses on building models from sample data in order to generate automated decision-making processes based what was originally analyzed. In simple words, machine learning takes an input of empirical data and predicts features of the data. Spine surgery research can benefit from a variety of machine learning approaches for supervised and unsupervised pattern analysis. In supervised learning the models are trained based on already available data and are typically used to make predictions on new data. In such a case, a mathematical function will map the input data pairs into output labels so that it can predict future outputs for new inputs (Fig. 1). On the other hand, unsupervised learning is the process of figuring out intrinsic structures and patterns of the data without any preset label given to the data. This means that the outcome is ignored when analyzing the data and the purpose of the analysis is to find hiding patterns and groupings in the data. Clustering is the most common computational tool used in unsupervised learning and it is widely used in genetic analysis (Fig. 1). For example, genetic clustering derived from molecular data showed that spinal ependymomas have a distinct genetic profile from their intracranial counterparts; an interesting finding to elucidate the mechanism of tumor formation [19].
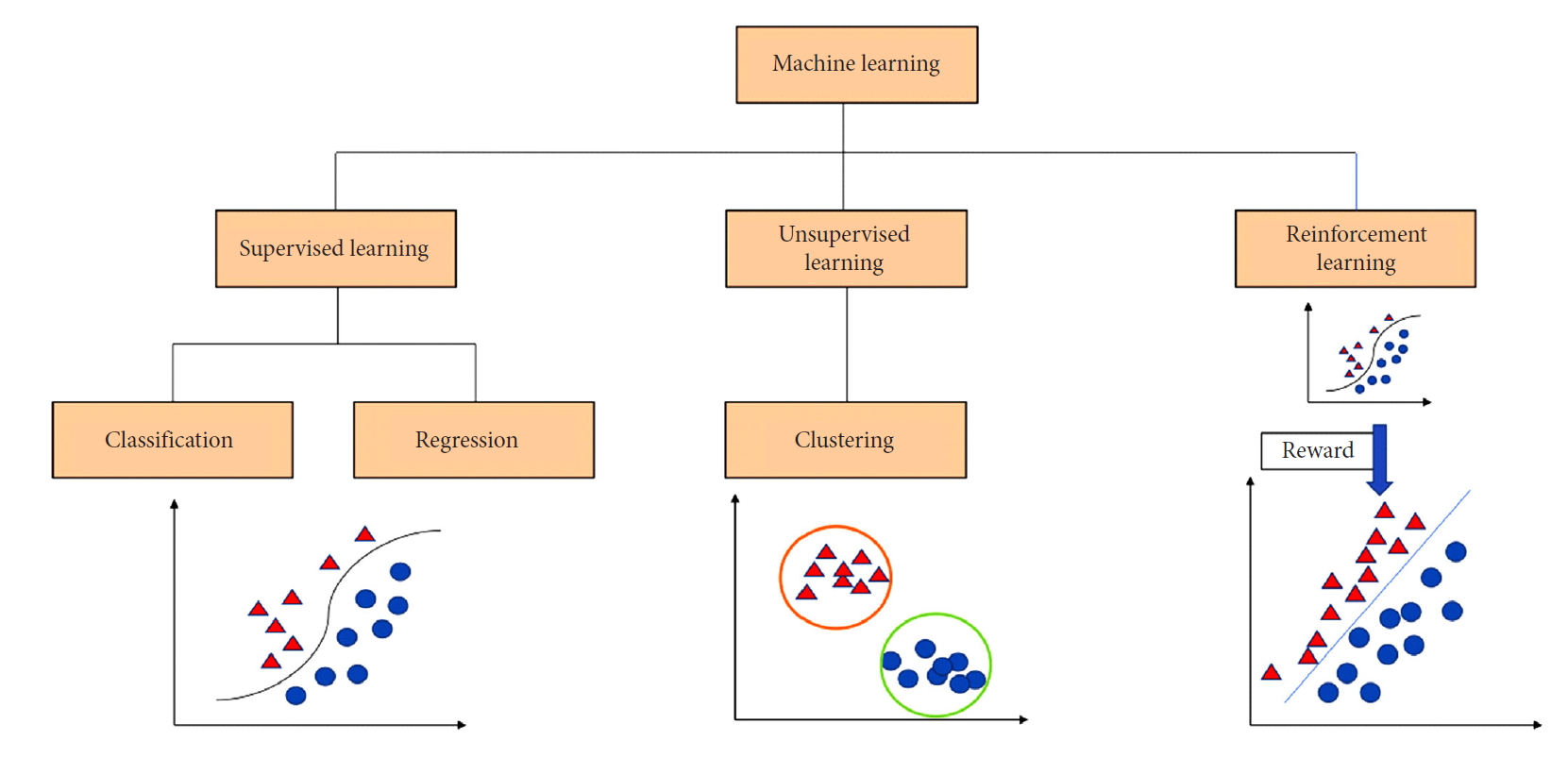
A classification chart that presents the 3 major branches of machine learning: (1) Supervised learning deals with classification of data and regression analysis to build a relationship between classes of inputs and outputs, (2) unsupervised learning and data clustering groups data irrespective of the output, and (3) reinforcement learning incorporates data to be able to select the best option from a pool of other options based on feedback and reward.
Reinforcement learning, which is another machine learning approach, is commonly used in dynamic environments where the agent is always in interaction with its environment to maximize rewards and minimize punishments or losses for a goal to optimize the outcome (Fig. 1). The concept of reinforcement learning gained attention when Google DeepMind applied it to Atari video games [20]. In this instance, the neural networks can be trained to maximize the game score by studying video game screen actions in order to maximize the likelihood of making the best next move. In the context of healthcare, reinforcement learning has been explored to optimize retroviral therapy for human immunodeficiency virus and determine the best approach for the step-wise management of sepsis [21,22]. These examples show how reinforcement learning can be used to solve sequential decision-making problems by extracting implicit knowledge from a big amount of data.
Spine researchers should also be familiar with the concepts of overfitting and underfitting models. The prediction model should always be balanced between optimization and generalization. In other words, the model should not be over adjusted to get the best performance possible on the training data, but most importantly should be adjusted and tested on data it has never seen before in order to claim generalization. When the performance of the model on the validation data begins to degrade, this means that the model is overfitting the original data by incorporating unusual features that are specific to the training data and therefore, are not generalizable. On the other hand, underfitting refers to a model that is not able to extract all relevant patterns in the training data, and therefore has not achieved either optimization of the training data or generalization for new data. The best way to avoid overfitting or underfitting of the data is to train the model on more data.
OVERVIEW OF CURRENT PREDICTION MODELS IN SPINE ONCOLOGY
Prediction models are the most common tools derived from big data analytics in healthcare. Prediction models for spinal metastases are designed to help in patient risk stratification and in decision making [8-10,12,23-27]. This is relevant as metastatic spinal cord compression occurs in up to 5% of all patients with cancer, most commonly in breast, lung, and prostate [28].
The number of patients included in the training sets of spinal metastasis prognostic scores ranges from n=61 (Tomita score, 2001) to n=1,790 (Skeletal Oncology Research Group machine learning algorithm, 2019) (Fig. 2). The study design of the different prognostic scores is summarized in Fig. 2 [8-10,12,23,24,26,27]. Early prognostic scores were developed mainly from retrospective studies conducted at single institutions, whereas more recent scores were developed using national databases (National Surgical Quality Improvement Program). It is evident that recent models have incorporated larger sample sizes, but the quality of the data is still mainly derived from retrospective analysis. Systematic reviews and meta-analysis studies looking for prognostic factors and outcomes in spinal metastatic disease include retrospective studies that are characterized by small sample sizes, selection bias and inaccuracy of the data reported. For example, survival outcomes are easy to calculate retrospectively but functional variables, which is a predictor of prognosis, may not be available at the time points desired in retrospective studies [29-32].

Commonly used prognostic scores for spine metastatic disease. For each model, the number of patients included, the method of data collection, the most common type of tumors included in the analysis, and the statistical tools used for building each model are listed.
Few prospective studies have been conducted to study surgical outcomes [31]. This speaks to the challenge of orchestrating such massive prospective data collection efforts. This can be very resource intensive and requires active data management and oversight. Schoenfeld et al. [33] recently proposed a study design for a prospective observational study for spinal metastasis treatment to validate the New England Spinal Metastasis score and accurately predict survival, morbidity, and functional outcomes in patients with spinal metastatic disease. Several other registered prospective studies, such as the EPOSO (Epidemiology, Process, and Outcomes of Spine Oncology Study) (NCT01825161), MTRON (Metastatic Tumor Research and Outcomes Network) study (NCT02830451), and PTRON (Primary Tumor Research and Outcome Network) study (NCT02790983) aim to collect demographic, medical, oncologic, radiographic, pathologic, clinical, diagnostic, and survival data. With such ongoing efforts, there will be opportunities for AI and machine learning applications from prospective data.
PERFORMANCE OF CURRENT MODELS
Many tools are used in biostatistics to assess the performance of prediction models. The c-statistic is considered a standard tool to test for discrimination. It tests the ability of a model to distinguish between 2 entities with respect to the outcome of interest. It is represented by the area under the curve (AUC) of receiver operating curves. A value of 1 indicates perfect discrimination and perfect model performance. A value of 0.5 indicates that the model is not better than flipping a coin at predicting the outcome. Usually, an AUC greater than 0.7 indicates a fair performance, and an AUC greater than 0.8 indicates a good performance.
Cox regression and logistic regression models have been used for developing most of the available prediction models and prognostic scores (Fig. 2) [8-12,23,24,27]. Cox regression is a statistical method used for survival analysis, whereas logistic regression is used to predict categorical outcomes. Logistic regression makes more assumptions about the data which introduces a higher risk of bias. This leads us to the variance-bias tradeoff: in general, more complex models have a high variance which may lead to overfitting. On the other hand, simple models tend to underfit if they are not flexible enough to model the true relationship and so have a high bias [34].
MACHINE LEARNING FOR PRIMARY TUMORS
Primary spine tumors (PST) are rare tumors that affect the spinal column. Tumors such as chordoma, chondrosarcoma, giant cell tumor, and osteosarcoma are all treated differently yet because of the rare occurrence of these tumors, uniform treatment algorithms do not exist and varies across centers [35]. As such, the reported data demonstrating clinical efficacy of various treatment strategies is often center dependent, subject to the inherent biases of the specific institution and treatment team. One institution may have expertise in radiation and not surgery. The opposite also applies. Considering the variability of such resources, any individual institution’s protocol or data may not be reproducible. So, the question remains: how to study rare tumors and predict outcomes when the collective intellectual experience is biased by local resources? The major limitation for building prediction models using machine learning for primary tumors of the spine is patient volume. A machine learning model predicting 5-year survival in chordoma patients was developed using a large database from the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) database using n=265 patients. This model had a good c-statistic discrimination index (AUC=0.8) but has not been externally validated to check its performance in a dataset that was not used initially for building the model.
Also using the same SEER database, a Chinese group successfully developed a nomogram to predict survival of Ewing’s sarcoma patients [36]. They used a validation and training dataset and achieved a fair performance (c-index=0.75). Other than survival, researchers are also working on state-of-the-art proteomics analyses using AI to efficiently deliver novel drugs for chordoma [37]. AI may enable the prediction of in vivo drug pharmacokinetics, pharmacodynamics, proper dosing, delivery methods, and platform design by using machine learning techniques.
PREDICTIVE ANALYTICS AND MOLECULAR MARKERS FOR SPINE TUMORS
Emerging studies are investigating the prognostic significance of tumor genomic and molecular biomarkers on therapies and clinical outcomes. At present, very few studies have attempted to integrate genomic and clinical biomarkers in predictive models [38,39]. To date, tumor histology has been the most commonly used factor in prediction models for spine metastatic disease but now with molecular-based therapies and immunotherapy, cancers need to be stratified further based on these molecular markers [40]. More recent systems such as the revised Katagiri have incorporated the molecular characteristics of the cancer in the prognostic model such as the hormone status of breast cancer and the molecular targets of non-small cell lung carcinoma [11]. We previously reported that lung cancer patients with identified oncologic molecular alterations have prolonged overall survival compared to patients with non-identified oncologic mutations [38]. Similarly outcomes for patients who received immunotherapy for metastatic melanoma has been shown to be worse after surgery despite longer overall cancer survival [41].
The prediction of clinical outcomes by taking into consideration genetic or molecular expression will be the next frontier step in predictive analytics [42]. Future approaches should consider the informational complexity of potential markers in relation to their biological environment. This will reveal the multivariate characteristics of cellular networks implicated in cancer progression. The AO Spine tumor group has developed the largest international collection of primary spine tumor samples with corresponding clinical data (n=1,495 samples) for genetic and survival analysis [43]. Bettegowda et al. [44] reported that human telomerase reverse transcriptase (hTERT) promotor mutations in spine chordoma confer a survival benefit and are potential prognostic molecular markers in spinal chordoma. Efforts continue to extract valuable information from spine tumor biobanks and link the available biologic data with prospective clinical data.
RADIOSURGERY FOR SPINE TUMORS
Radiosurgery has significantly changed the treatment of spinal metastasis and PST with its precision, safety, and outstanding efficacy [45]. Conventional prediction models and prognostic scores are limited as they do not take into consideration newer treatment modalities such as stereotactic radiosurgery [8,9,24,27].
Data demonstrate that radiosurgery yields a clinical benefit regardless of tumor histology and volume, providing durable symptomatic responses and high local-control rates [46]. In many situations, radiosurgery has largely replaced en bloc resection as favored by the Tokuhashi and Tomita scoring systems [47,48]. A study group at Texas MD Anderson developed a Prognostic Index for Spine Metastasis to risk-stratify patients who receive radiosurgery treatment for spinal metastatic disease [49]. This scoring system is composed of gender, Karnofsky Performance Status, previous surgery at the site of radiosurgery, previous radiation at the site of radiosurgery, the extent of disease metastasis, solitary metastasis, and time from diagnosis to metastasis [49]. Another study identified that radioresistant tumors including nonsmall cell lung carcinoma and colorectal carcinoma have a propensity toward local failure after treatment with radiosurgery [50].
Machine learning and deep learning methods have been recently used for auto-segmentation and contouring of tumors in head and neck oncology. Use of this technology saves clinicians time and produces good quality radiation planning contours [51-53]. The same technology can potentially be used for spine tumors to increase radiation treatment accuracy, target delineation, and standardize protocols among institutions.
CONCLUSION
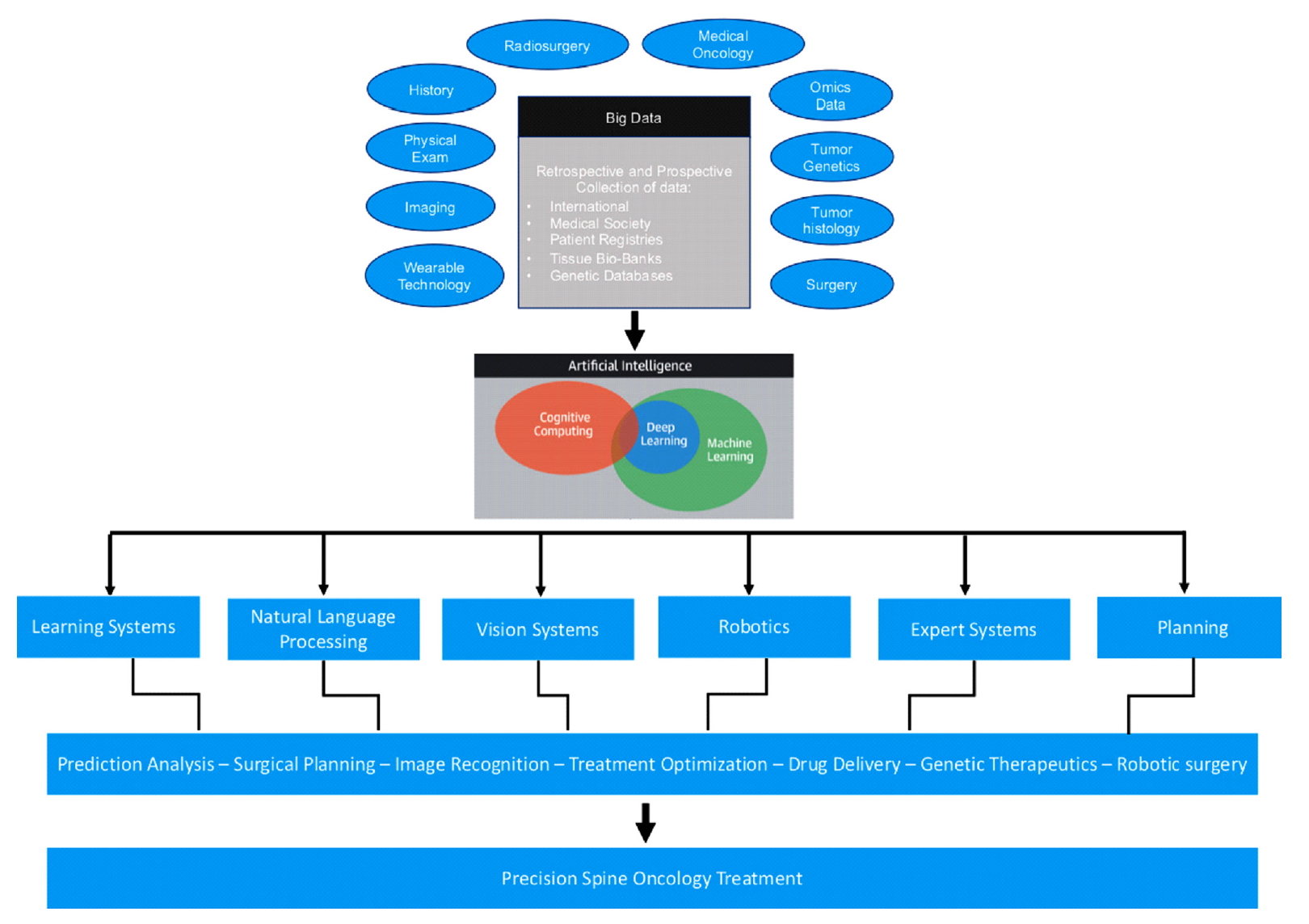
The field of AI is constantly expanding, driven by advances in technology and computer science. Data continues to be the most crucial component for learning AI systems and applying such systems to the field of spine oncology is in its infancy. With the numerous data points associated with each clinical scenario, AI will help drive the field forward with more precise and unbiased assessments of survival and effectiveness of interventions (Fig. 3). Future predictive algorithms in spine oncology will need to incorporate radiographic, pathologic, molecular, and oncologic data in a more rigorous fashion. The challenge is identifying and assigning weights to the vast number of data points in the medical record and inputting that data in real time. The application of AI to spine oncology can hopefully help develop better data-driven clinical decision-making tools for more precise care.
Notes
The authors have nothing to disclose.