 |
 |
- Search
| Neurospine > Volume 21(1); 2024 > Article |
|
|
Abstract
Objective
Computed tomography (CT) imaging is a cornerstone in the assessment of patients with spinal trauma and in the planning of spinal interventions. However, CT studies are associated with logistical problems, acquisition costs, and radiation exposure. In this proof-of-concept study, the feasibility of generating synthetic spinal CT images using biplanar radiographs was explored. This could expand the potential applications of x-ray machines pre-, post-, and even intraoperatively.
Methods
A cohort of 209 patients who underwent spinal CT imaging from the VerSe2020 dataset was used to train the algorithm. The model was subsequently evaluated using an internal and external validation set containing 55 from the VerSe2020 dataset and a subset of 56 images from the CTSpine1K dataset, respectively. Digitally reconstructed radiographs served as input for training and evaluation of the 2-dimensional (2D)-to-3-dimentional (3D) generative adversarial model. Model performance was assessed using peak signal to noise ratio (PSNR), structural similarity index (SSIM), and cosine similarity (CS).
Results
At external validation, the developed model achieved a PSNR of 21.139 ± 1.018 dB (mean ± standard deviation). The SSIM and CS amounted to 0.947 ± 0.010 and 0.671 ± 0.691, respectively.
Conclusion
Generating an artificial 3D output from 2D imaging is challenging, especially for spinal imaging, where x-rays are known to deliver insufficient information frequently. Although the synthetic CT scans derived from our model do not perfectly match their ground truth CT, our proof-of-concept study warrants further exploration of the potential of this technology.
Computed tomography (CT) imaging is an essential modality in the diagnosis of neurosurgical disease [1-3]. In spine surgery, is routinely employed to assess the extent of injury or signs of degeneration [1,4]. Furthermore, CT imaging plays a vital role in surgical intervention planning for spinal tumors, degenerative as well as infectious spinal diseases, and can be required for establishing a precise neuronavigation setup [5,6].
Despite advances in radiation technology, patients remain exposed to significant doses of ionizing radiation. While the average annual background radiation in the United States is estimated at approximately 3 mSv, the radiation exposure for a lumbar spine CT scan are much lower, typically around 1.5 mSv [7]. However, in cases of polytrauma, extensive examination is indicated so that radiation doses can exceed 20 mSv [8].
Patients with severe spinal cord injuries may also be hemodynamically unstable and require immobilization, making CT imaging challenging [9,10]. Moreover, due to financial constraints, many centers worldwide have limited access to this specialized modality [11]. To maintain full operational capability, radiographers and radiologists are necessary, further adding to the financial burden.
Generating 3-dimensional (3D) information from a 2-dimensional (2D) input has the potential to assist in addressing these issues. The feasibility of creating synthetic, CT-like 3D imaging using deep learning from biplanar chest x-ray images has been demonstrated [12]. A similar machine learning based application for spine imaging, however, is currently missing. In this proof-of-concept study, the main objective was to assess the feasibility of creating synthetic spinal CT images using deep learning by using biplanar digitally reconstructed radiographs (DRRs) as inputs, serving as a proxy for true biplanar x-ray images.
Noncontrast enhanced spinal CT scans from 209 patients and their DRRs, sourced from the VerSe2020 dataset, were used for model development [13-15]. The VerSe20 dataset also includes manually refined segmentations of the vertebrae. To generate 2D-to-3D synthetic CT (sCT) images, a generative adversarial network (GAN) model was trained on these images, following the methodology previously established by Ying et al. [12]. Model evaluation was subsequently carried out on 55 cases from the VerSe20 dataset [13-15] (internal validation) and a randomly selected subset of 56 patients from the CTSpine1K dataset [16] (external validation) using structural similarity index (SSIM), peak signal to noise ratio (PSNR), and cosine similarity (CS) as metrics.
The VerSe20 and CTSpine1K datasets are publicly available and ethical approval has been obtained by the original publishers from the corresponding ethics committees.
The CT data used for model development and evaluation originated from multiple centers and was obtained from the VerSe2020 dataset. This publicly available dataset includes spinal imaging from cervical, thoracic and lumbar regions acquired on scanners from multiple manufacturers and features a variety of pathologies, such as vertebral fractures, deformities, and implants [13-15]. For external validation a randomly selected subset of 56 images originating from the MSD-T10 and COVID-19 datasets, which are both part of the CTSpine1K dataset [16], was applied. Some further information on the datasets is provided in Table 1.
Given the necessity for corresponding radiographs to the CT scans, in this feasibility study, DRRs were derived—serving as a proxy for true biplanar x-rays. They were generated from the 3D-CT scans using the Plastimatch software [17], which offers relatively high fidelity to actual x-ray images, making them an ideal basis for model training. However, it is important to note that DRRs are not exact replicas of conventional x-rays. In order to highlight bone structures, Hounsfield units (HU) windowing was applied with a window center of 400 HU and a window width of 1,300 HU. In addition, the vertebrae segmentations were employed to extract the bones. The pelvis and sacrum were excluded from consideration.
For assessment of model performance, 3 evaluation metrics were employed: PSNR, SSIM, and CS. PSNR serves as a widely accepted quantitative metric for determining the fidelity of reconstructed digital signals, with higher values indicating superior image quality [18]. SSIM provides a multifaceted assessment of image similarity by considering factors like luminance, contrast, and structure [18,19]. Unlike metrics such as mean squared error and PSNR that focus on absolute errors only, SSIM is designed to correspond more closely with human perception of image quality. Finally, CS is a can be applied to measure similarity by assessing the cosine of the angle between 2 feature vectors [20]. Results for both SSIM and CS range from -1 (completely dissimilar) to 1 (identical). As PSNR values depend on the bit depth of the image, there is no fixed definition of a good score [18].
SSIM is calculated once for all 3 dimensions and averaged thereafter. In order to calculate CS images were flattened into a 1-dimensional vector.
All CT scans were resampled to achieve a uniform voxel spacing of 1× 1× 1 mm³ and the image dimensions were standardized to 256× 256× 256 pixels. Thereafter, biplanar DRRs were derived for all CT scans. These DRRs acted as the input for both training and assessing our GAN model. The CT images were then windowed, and the spinal structures extracted as described before. Finally, the total number of unique grayscale values was compressed to 100, while maintaining the integrity of the overall data distribution. This was accomplished through the development of a lookup table, which enabled a systematic mapping of HU to their corresponding bins (quantization).
A GAN was fine-tuned on the training set, allowing us to create sCTs from biplanar DRRs. The basic model architecture was adapted from X2CT-GAN [12], and certain parameters were adjusted to meet the specific requirements of our task. The final model underwent 100 epochs of training with a learning rate of 0.00005. Final evaluation was carried out on the 2 holdout sets to assess PSNR, SSIM, and CS. The sCTs were generated to have a resolution of 192× 192× 192 voxels. Fig. 1 schematically depicts the data extraction, model application and evaluation process. All calculations were performed with Python 3.10.6 and PyTorch 1.12.1 for CUDA 11.6.
Table 2 outlines model performance. A visual comparison between synthetic images generated by our GAN and the corresponding ground truth data is depicted in Fig. 2 for a case taken from the internal validation and Fig. 3 from the external validation set.
Fig. 4 depicts a comparison in 3D space of the synthetically generated spinal column derived from DRRs and the ground truth version obtained through manual segmentation.
Utilizing a multicenter dataset encompassing over 300 patients, a 2D-to-3D GAN capable of synthesizing spinal CT images from biplanar radiographs was successfully developed and externally evaluated. In terms of performance, the model yielded promising but not fully satisfying results, revealing that while the sCT are not able to completely replace traditional CT scans, they might offer a viable alternative in certain clinical settings.
The generation of sCT images from x-rays holds promise for reducing time, cost, and radiation exposure associated with CT scans. Unlike CT, radiographic imaging has major limitations to performing 3D anatomical evaluations. This study proposes a model capable of generating sCTs using DRRs as input. While these synthetically generated images may not match the quality of actual CT scans in every respect, it is possible that a more advanced version of the model, trained on a larger dataset, at higher resolution and on real radiographs instead of DRRs, could provide sufficient diagnostic information to make a real CT redundant in specific scenarios. The aim should not be to replace CTs as a whole, but much rather extend the current applications of radiography machines. We believe that by using x-rays from 2 planes (anterior-posterior and lateral), it should be possible to obtain enough depth information to generate reliable, 3D images using deep learning. The depth information and consequently the quality of our synthetic imaging will absolutely be limited compared to authentic CTs. Such an application, however, would for example be very beneficial in low resource settings, where CTs are not accessible but x-ray machines are.
Apart from reducing radiation exposure and improving economic efficiency, our model could also directly influence patient treatment. For instance, fluoroscopy machines are commonly used in neurosurgical operating theatres for spinal surgeries. By using such fluoroscopic imaging as input, 3D imaging could be generated, thus enhancing orientation and decreasing spatial awareness problems during surgery.
The model training approach we used necessitates manual vertebrae segmentation. We discovered that omitting the bone extraction leads to less uniform results in the generated sCTs. This lack in performance is likely due to the variable characteristics of the surrounding soft tissue, which appeared to interfere with the model’s ability to learn appropriately. Such a manual approach dramatically increases the training time and introduces the potential for human error. Additional work, on a new automated deep-learning segmentation model to streamline the training process is ongoing. However, it needs to be reiterated, that although the model has been trained semiautomatically, its application on new data is already fully automated, as all that is required for the generation of the sCT are the biplanar radiographs/DRRs.
Quantization was employed in order to reduce the number of individual grayscale images in our CTs while retaining as much of the overall data distribution as possible. This significantly decreases computational costs of model training with the drawback of losing the exact Hounsfield intensities and loosing image depth. The exact number of gray shades a human eye can discern is debated, however medical displays typically offer 8 bits or 256 shades of gray [21]. In such scenarios, the benefits of higher-depth imaging can only be taken advantage of by adjusting contrast and brightness. By using a binning approach with 100 bins (grayscale units), our method remains below this threshold. However, we plan to explore expanding the grayscale range in the future.
It is important to exercise caution when interpreting CS as a measure for image similarity. While PSNR and SSIM are established metrics for image fidelity assessment, to the best of our knowledge, there is no literature available that investigates CS’s utility for this purpose. CS is a commonly used similarity measure applied to text documents, for example for clustering [22]. As, based on our personal observation, this metric corresponded relatively well with our subjective qualitative assessment of the synthetic imaging, we decided on including it in our evaluation anyway.
Our model performance in terms of SSIM appear quite high, reaching 0.947 at external validation. However, it is important to note that, as we remove all the soft tissue from our imaging, a substantial part of our synthetic and ground truth CTs is made up of background. As SSIM is calculated over the entire image and the spinal column only constitutes a comparably small fraction of the overall volume, the model easily reconstructs the background which results in a high SSIM score.
There have been multiple approaches aimed at generating higher-dimensional images from single-plane radiographs [23,24]. However, these approaches lack depth information due to their reliance on a single view so that the generated images may not offer the accuracy needed for precise diagnostic or surgical planning. In contrast, deep-learning approaches using multiplane radiographs as input in order to create CT-like imaging have been successful [12,25]. Furthermore, several previous studies describe spine-specific methodologies to extrapolate 3D information from biplanar, 2D radiographic imaging [26-33]. Most of these studies employed statistical approaches, while only a smaller subset has leveraged machine learning. Notably, instead of generating CT-like imaging, all these investigations focused on generating 3D models of the spine. To the best of our knowledge, only one previous study attempted to apply a GAN in a similar fashion for generating synthetic, CT-like spinal images from biplanar DRRs [34]. Saravi et al. [34], who independently performed their study in parallel to ours, used DRRs generated after extraction of the vertebrae for the generation of the sCTs, while we derived the DRRs before extraction. Therefore, the DRRs used to train our model are more similar to authentic x-rays. The major next step towards clinical applicability, however, will be applying this technique to real x-rays or fluoroscopy images, instead of DRRs.
To translate a model like the one we propose into clinical practice, several steps are required. First, the model needs to be thoroughly evaluated in silico. To achieve this, it needs to be trained on more data originating from as many centers as possible, while additionally increasing data variability by applying data augmentation methods. Future research should also assess the performance of the model separately for axial and sagittal 2D image slices. These 2 planes are of especially high diagnostic relevance and axial imaging can only be reconstructed indirectly when using anterior-posterior and lateral x-rays as input, presumably making it more error-prone. Furthermore, future studies need to perform a more accurate assessment by using real radiograph images as input instead of DDRs. Once this has been established, clinical trials to assess the model’s utility in clinical practice are necessary. This could be accomplished by comparing authentic CT scans to the synthetic imaging for specific inquiries such as implant placement after spinal fusion surgery or more general diagnostic questions. Only after conducting clinical trials can the true clinical utility of a model, like the one we propose, really be understood.
Even though the developed model was trained on a rather large and diverse training dataset, encompassing 209 spinal images that spanned the lumbar, thoracic, and cervical regions, more data is likely to improve its performance. This is particularly relevant for spinal imaging as it can exhibit significant variability between individuals.
Notably, cervical imaging was underrepresented in the training and evaluation data, and model performance based on spinal region needs to be evaluated in future studies. Similarly, the impact of surgical implants on our model’s performance has not been assessed yet.
Although established image quality metrics (PSNR, SSIM, and CS) were employed, these measurements probably do not capture the full clinical utility of the generated sCTs. For a detailed understanding of the model’s applicability, additional clinically relevant metrics need to be included in combination with an expert radiological evaluation.
In this proof-of-concept study, we managed to successfully externally validate our model. However, to ensure adequate generalizability, validation on data from further centers is would be beneficial.
Lastly, DRRs were used as training input as a substitute for real x-rays, being an initial proof-of-concept study. DRRs offer high fidelity and serve as a useful approximation but they cannot fully replace conventional x-rays. One of the major challenges we faced in using real x-rays is the lack of correspondence of x-rays to CTs in terms of orientation and relative size. Frequently, the x-ray beam is not perpendicular to the skull, and the distance from the x-ray source can also vary. It is thus essential for future research to explore methods that allow for the use of authentic x-rays as model input.
We present a proof-of-concept study of TomoRay, a GAN targeted at generating synthetic CT scans of the cervical, thoracic, and lumbar spine from biplanar imaging. While the quality of our algorithm-generated CT scans has not yet reached the level of authentic CT scans, our study warrants further exploration of the potential of this technology.
NOTES
Fig. 1.
Data extraction, model application and evaluation process. (A) Application of mask and Hounsfield windowing in order to obtain ground truth computed tomography (CT) for algorithm training. (B) Generation of digitally reconstructed radiographs (DRRs) using Plastimatch software. (C) Application of the fully trained model to new data and evaluation. 2D, 2-dimensional; 3D, 3-dimensional; GAN, generative adversarial network.

Fig. 2.
Example of synthetic computed tomography (CT) generated by our model on the internal validation set. For panels A and B from left to right: ground truth CT, manually extracted vertebra from ground truth CT, synthetic CT generated by our model. (A) Axial view. (B) Sagittal view. (C) Digitally reconstructed radiograph derived from the ground truth CT and used as model input.
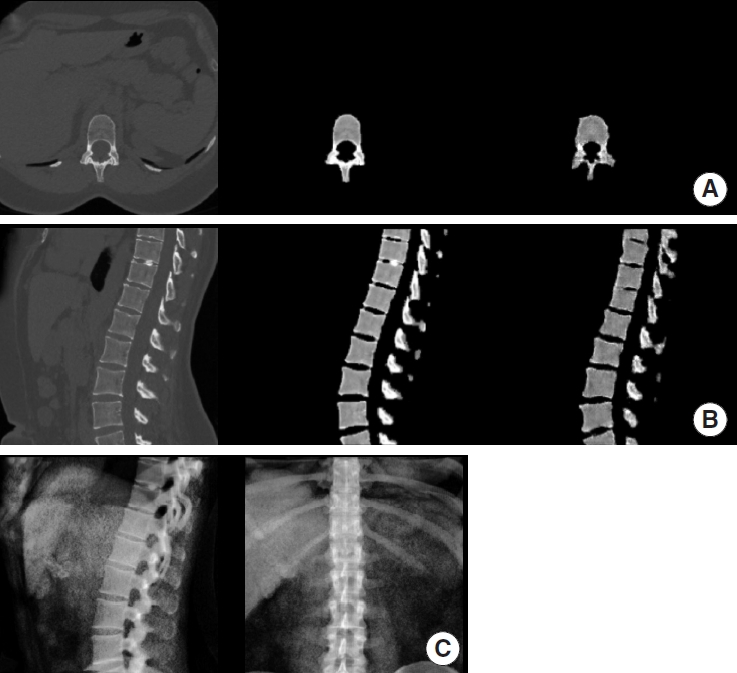
Fig. 3.
Example of synthetic computed tomography (CT) generated by our model on the external validation set. For A and B from left to right: ground truth CT, manually extracted vertebra from ground truth CT, synthetic CT generated by our model. (A) Axial view. (B) Sagittal view. (C) Digitally reconstructed radiograph derived from the ground truth CT and used as model input.
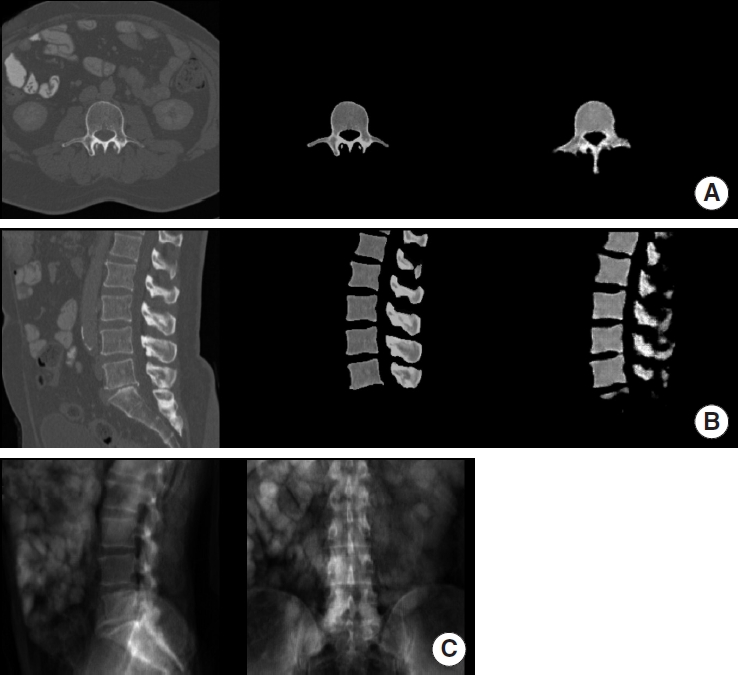
Fig. 4.
Reconstruction of the synthetic computed tomography generated by the model in 3-dimensional space (case from the internal validation set). (A) Ground truth. (B) Model generated.

Table 1.
Overview of the dataset partition used in this study
Table 2.
Model performance on training and holdout sets
SSIM is calculated for each dimension individually and then averaged for each image. The final values reported are the mean and median over the whole dataset.
PSNR, peak signal to noise ratio; SSIM, structural similarity index; 3D, 3-dimensional; 2D, 2-dimensional; SD, standard deviation; IQR, interquartile range.
REFERENCES
1. Holmes JF, Akkinepalli R. Computed tomography versus plain radiography to screen for cervical spine injury: a meta-analysis. J Trauma Acute Care Surg 2005;58:902-5.

2. Kirkman MA. The role of imaging in the development of neurosurgery. J Clin Neurosci 2015;22:55-61.


3. Rodallec MH, Feydy A, Larousserie F, et al. Diagnostic imaging of solitary tumors of the spine: what to do and say. RadioGraphics 2008;28:1019-41.


4. van Den Hauwe L, Sundgren PC, Flanders AE. Spinal trauma and spinal cord injury (SCI). 2020 Feb 15. In: Hodler J, Kubik-Huch RA, von Schulthess GKet al., editors. Diseases of the brain, head and neck, spine 2020–2023: Diagnostic Imaging [Internet]. Cham (CH): Springer; 2020.Chapter 19.
5. Haberland N, Ebmeier K, Hliscs R, et al. Neuronavigation in surgery of intracranial and spinal tumors. J Cancer Res Clin Oncol 2000;126:529-41.


6. Moses ZB, Mayer RR, Strickland BA, et al. Neuronavigation in minimally invasive spine surgery. Neurosurg Focus 2013;35:E12.

7. McCollough CH, Bushberg JT, Fletcher JG, et al. Answers to common questions about the use and safety of CT scans. Mayo Clin Proc 2015;90:1380-92.


8. Tien HC, Tremblay LN, Rizoli SB, et al. Radiation exposure from diagnostic imaging in severely injured trauma patients. J Trauma 2007;62:151-6.


9. Galeiras Vázquez R, Rascado Sedes P, Mourelo Fariña M, et al. Respiratory management in the patient with spinal cord injury. BioMed Res Int 2013;2013:168757.


11. Bergeron C, Fleet R, Tounkara FK, et al. Lack of CT scanner in a rural emergency department increases inter-facility transfers: a pilot study. BMC Res Notes 2017;10:772.



12. Ying X, Guo H, Ma K, et al. X2CT-GAN: reconstructing CT from biplanar X-Rays with generative adversarial networks. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019;10611-20.

13. Liebl H, Schinz D, Sekuboyina A, et al. A computed tomography vertebral segmentation dataset with anatomical variations and multi-vendor scanner data. Sci Data 2021;8:284.



14. Löffler MT, Sekuboyina A, Jacob A, et al. A vertebral segmentation dataset with fracture grading. Radiol Artif Intell 2020;2:e190138.



15. Sekuboyina A, Husseini ME, Bayat A, et al. VerSe: a vertebrae labelling and segmentation benchmark for multi-detector CT images. Med Image Anal 2021;73:102166.


16. Deng Y, Wang C, Hui Y, et al. CTSpine1K: a large-scale dataset for spinal vertebrae segmentation in computed tomography [Internet]. arXiv:2105.14711v3 [Preprint]. 2021 [cited 2023 Dec 12]; Available at: http://arxiv.org/abs/2105.14711.
17. Sharp GC, Li R, Wolfgang J, et al. Proceedings of the XVI’th International Conference on the use of Computers in Radiotherapy (ICCR). Amsterdam: Netherlands; 2010.
18. Sara U, Akter M, Uddin MS. Image quality assessment through FSIM, SSIM, MSE and PSNR—a comparative study. J Comput Commun 2019;7:8-18.

19. Renieblas GP, Nogués AT, González AM, et al. Structural similarity index family for image quality assessment in radiological images. J Med Imaging 2017;4:035501.



20. Xia P, Zhang L, Li F. Learning similarity with cosine similarity ensemble. Inf Sci 2015;307:39-52.

21. Kimpe T, Tuytschaever T. Increasing the number of gray shades in medical display systems—how much is enough? J Digit Imaging 2007;20:422-32.

22. Strehl A, Ghosh J, Mooney R. Impact of similarity measures on web-page clustering. Workshop on artificial intelligence for web search (AAAI 2000). Washington, DC: Association for the Advancement of Artificial Intelligence; 2000. p. 64.
23. Loÿen E, Dasnoy-Sumell D, Macq B. Patient-specific three-dimensional image reconstruction from a single X-ray projection using a convolutional neural network for on-line radiotherapy applications. Phys Imaging Radiat Oncol 2023;26:100444.



24. Lu S, Li S, Wang Y. Prior information-based high-resolution tomography image reconstruction from a single digitally reconstructed radiograph. Phys Med Biol 2022;67(8):doi: 10.1088/1361-6560/ac508d.

25. Almeida DF, Astudillo P, Vandermeulen D. Three-dimensional image volumes from two-dimensional digitally reconstructed radiographs: a deep learning approach in lower limb CT scans. Med Phys 2021;48:2448-57.


26. Aubert B, Vazquez C, Cresson T, et al. Toward automated 3D spine reconstruction from biplanar radiographs using CNN for statistical spine model fitting. IEEE Trans Med Imaging 2019;38:2796-806.


27. Chen Z, Guo L, Zhang R, et al. BX2S-Net: learning to reconstruct 3D spinal structures from bi-planar X-ray images. Comput Biol Med 2023;154:106615.


28. Gille O, Champain N, Benchikh-El-Fegoun A, et al. Reliability of 3D reconstruction of the spine of mild scoliotic patients. Spine (Phila Pa 1976) 2007;32:568-73.


29. Glaser DA, Doan J, Newton PO. Comparison of 3-dimensional spinal reconstruction accuracy: biplanar radiographs with EOS versus computed tomography. Spine (Phila Pa 1976) 2012;37:1391-7.

30. Humbert L, De Guise JA, Aubert B, et al. 3D reconstruction of the spine from biplanar X-rays using parametric models based on transversal and longitudinal inferences. Med Eng Phys 2009;31:681-7.


31. Le Bras A, Laporte S, Mitton D, et al. Three-dimensional (3D) detailed reconstruction of human vertebrae from low-dose digital stereoradiography. Eur J Orthop Surg Traumatol 2003;13:57-62.

32. Zhang J, Lv L, Shi X, et al. 3-D reconstruction of the spine from biplanar radiographs based on contour matching using the hough transform. IEEE Trans Biomed Eng 2013;60:1954-64.



- TOOLS
-
METRICS

-
- 1 Crossref
- Scopus
- 1,024 View
- 58 Download
- Related articles in NS
-
Journal Impact Factor 3.2


























