- Search
|
|
||
See commentary "Commentary: Artificial Intelligence for Adult Spinal Deformity" in Volume 16 on page 695.
Abstract
Adult spinal deformity (ASD) is a complex disease that significantly affects the lives of many patients. Surgical correction has proven to be effective in achieving improvement of spinopelvic parameters as well as improving quality of life (QoL) for these patients. However, given the relatively high complication risk associated with ASD correction, it is of paramount importance to develop robust prognostic tools for predicting risk profile and outcomes. Historically, statistical models such as linear and logistic regression models were used to identify preoperative factors associated with postoperative outcomes. While these tools were useful for looking at simple associations, they represent generalizations across large populations, with little applicability to individual patients. More recently, predictive analytics utilizing artificial intelligence (AI) through machine learning for comprehensive processing of large amounts of data have become available for surgeons to implement. The use of these computational techniques has given surgeons the ability to leverage far more accurate and individualized predictive tools to better inform individual patients regarding predicted outcomes after ASD correction surgery. Applications range from predicting QoL measures to predicting the risk of major complications, hospital readmission, and reoperation rates. In addition, AI has been used to create a novel classification system for ASD patients, which will help surgeons identify distinct patient subpopulations with unique risk-benefit profiles. Overall, these tools will help surgeons tailor their clinical practice to address patientsŌĆÖ individual needs and create an opportunity for personalized medicine within spine surgery.
For decades, adult spinal deformity (ASD) has continued to be a complex and often crippling disease, causing significant pain and disability [1,2], with worse deformity associated with greater pain and disability [3,4]. ASD is not a single entity but a very heterogeneous disease with many etiologies: degenerative, idiopathic, congenital and commonly iatrogenic (prior surgery). While our understanding of the complexity of this disease has increased, so has our ability to surgically manage this condition, with many studies showing that correction of spinopelvic parameters to more normal values can significantly improve multiple health-related quality of life (HRQoL) measures especially in severely disabled patients [5-8]. In almost all cases, soft tissue release and osteotomies are required in order to obtain satisfactory correction of the deformity. In rigid, inflexible ASD cases, high grade 3-column osteotomies may be warranted [4]. Such formidable techniques are associated with greater surgical invasiveness, risk for complications (perioperative and long-term), neurological risks, and direct cost [7,9-14].
Due to the unique nature of ASD and multifaceted nature of the patients, ASD offers a perfect niche for utilization of advanced analytics throughout nonsurgical and surgical care. For decades, spine surgeons have relied on established literature, extensive training, and clinical judgment to counsel patients regarding the risks and benefits of surgery for ASD; often the most accurate information was based on their overall personal experience and was not patient specific. Most studies in the literature were conducted using simple statistical methods such as linear or logistic regressions, and gave surgeons averages across entire populations that in reality may be minimally relevant for the intricacies of a specific patient. As medical data has become digitized giving us access to dizzying arrays of patient information, so has our ability to process this data in far more meaningful and robust ways. The past few years have seen the medical fieldŌĆÖs gradual adoption of computational techniques that can process vast amounts of data to create complex mathematical models describing the relationship between seemingly disparate variables. The most widely used of these techniques has been machine learning, which has surged in popularity over the past decade as the most commonly utilized tool for implementing artificial intelligence (AI).
The idea behind AI, is to create a system that can mimic our natural ability to continually learn as we gain access to new data and apply it to novel situations in the future. In the context of machine learning, which is considered a subset of AI, this involves ŌĆ£trainingŌĆØ algorithms on immense datasets, and allowing the algorithm to discern mathematical relationships within the data (Fig. 1). Once an algorithm has been trained on previously acquired data, it can then prospectively be applied to new data in order to make specific predictions or determinations based on the model that has been developed. This ability to interpret patterns in data that may not be readily apparent offers powerful applications to the field of spine surgery, especially ASD. Given the wide spectrum of data available for patients undergoing ASD surgery, incorporating machine learning algorithms into predictive analytics helps remove bias regarding which variables are relevant or not, and has the potential to make predictions tailored specifically to a patientŌĆÖs individual profile. The applicability of machine learning models to prospective data for individual patients offers an immense advantage over traditional statistical methods that portray estimates for largely diverse patient populations with little prospective utility. As a result, machine learning models can meaningfully augment a surgeonŌĆÖs ability to counsel patients. Spinal deformity surgeons are at the forefront of incorporating these techniques for a wide variety of potential applications, including predictive modeling for outcomes, cost analysis for both patients and healthcare systems, and complication risk profiling.
For ASD specifically, spine surgeons have made great strides in the development and implementation of machine learning techniques. The largest advancements have been in the utilization of increasingly complex predictive analytics that is built on machine learning algorithms. Both surgeons and patients alike are interested in better tools for predicting outcomes, as it allows for more comprehensive preoperative discussions and surgical decision making. Predictive analytics has now been applied across a wide variety of topics within ASD surgery, including predicting intraoperative [15], perioperative [16,17], and postoperative complications and outcomes [18-25].
The majority of studies published on this topic share similar principles and methodologies in the development of their respective predictive models. The most common technique employed across the studies mentioned in this article relies on decision tree-based machine learning, where either classification or regression trees are built based on the target variable (output). The idea behind decision tree learning is that the algorithm will build a tree-like model of decisions and their corresponding consequences (similar to a flow chart) (Fig. 2). In this case, the tree will navigate from observations represented as the ŌĆ£branchesŌĆØ (clinical variables) about an item (the patient) to conclusions about the itemŌĆÖs target (desired output ŌĆō outcome variable) represented as the ŌĆ£leaves.ŌĆØ
In order to create the predictive model, these decision trees are first created by learning from a ŌĆ£training set,ŌĆØ which is generally a 70%ŌĆō80% partition of the entire dataset. Once created, the parameters of the model are fine-tuned using a ŌĆ£validation setŌĆØ, generally a 20%ŌĆō30% partition of the data. Final metrics for testing the accuracy of predictions generated by the predictive model are usually derived using a ŌĆ£test setŌĆØ where the actual outputs are already known, and reported using metrics such as % accuracy and area under the curve (AUC) (Fig. 3). Variations of the decision tree learning concept help make the analytics more robust and generalizable (minimize overfitting) for making predictions on new data. These variations include using bootstrapping, which randomly samples data for the creation of decision trees during the training phase, and random forest algorithms ŌĆō a slight modification of the actual decision tree algorithm that randomly selects a subset of features (variables) and builds decision trees with different structures to find variables which are the strongest predictors of the desired output. Ensemble methods such as random forests or bootstrapped decision trees combine several different learned algorithms (differently structured trees) to create a single stronger classifier that will have better predictive value and lower variance.
While statistical models still remain highly relevant for healthcare analytics, they do possess several limitations in their applicability, especially when compared to machine learning. The primary differences between statistical modeling and machine learning lie in their data requirements, ease of interpretation and understanding of the model generated. Statistical modeling serves the purpose of actually explaining or inferring the relationship between variables in a model. The strength of machine learning, on the other hand, lies in its capability to process immense amounts of data across large numbers of often diverse variables, to generate predictions with high accuracy for specific outcomes. The tradeoff here is that statistical models while less accurate for predicting outcomes are generally easier to interpret, and machine learning models which grant higher predictive accuracy are more difficult to interpret given their increased complexity.
Machine learning algorithms are extremely powerful when used appropriately but present some limitations, especially regarding their adoption as an analytic tool for medicine. A key difference between machine learning methods and statistical modeling lies with their differing requirements for data. In general, statistics can be applied to relatively small amounts of data and still allow for reasonable inferences to be made of the relationship between variables. Machine learning, on the other hand, requires much larger amounts of data in order to effectively create predictive models, which then improve with subsequent addition of new data. Given the scarcity of large prospectively collected datasets in spine surgery, some of the predictive models described later on should be viewed cautiously, as predictive accuracy can vary greatly when sufficient data is not provided to train machine learning models. Additionally, given the readily accessible applications that currently exist for applying machine learning, many of the subtleties underlying the implementation of these models are often lost to the general user. Some of these include being careful about managing data that exists in different forms (comorbidities, lab results, binary outcomes, free text, etc.), and skimping on model and parameter training.
Oftentimes in medicine, we are presented with class imbalance problems, where one outcome class can represent a significant majority and outweigh a different minority outcome, causing our predictions to become heavily biased towards one way or another. Additionally, models that are not trained on sufficient sample sizes can be hindered by overfitting, meaning the model is effective at describing the existing data, but cannot be extrapolated with the same accuracy to new data. Data scientists utilize many common techniques to circumvent these shortcomings, most common of which requires proper training, validation, and testing of the desired model. Physicians and researchers must take care to follow appropriate steps in development of machine learning models for clinical outcomes research, as avoidance of such practices as well as careful parameter calibration and tuning can result in misleading conclusions.
Though predictive analytics have primarily been applied to postoperative outcomes, Durand et al. [15] studied a group of 1,029 ASD patients to develop a predictive model for intra- and postoperative blood transfusion. Single decision tree and random forest models were both developed with a training set of 824 patients and tested on a validation set of 205 patients. The final classification tree model and random forest model exhibited AUCs of 0.79 and 0.85, respectively, with no significant difference between the 2 models. The resulting models can provide surgeons with accurate tools to predict transfusion rates among their ASD patients, allowing for more informed surgical planning. Models have also been created to assess length of stay (LOS) [16] and major early complications [17]. In the study by Scheer et al. [17] a predictive model for early complications (intraoperative and within 6 weeks postoperative) was created using 45 variables from baseline demographic, radiographic, and surgical factors for 557 ASD patients. An ensemble of decision trees was trained with 5 different bootstrapped models and internal validation was accomplished using a 70:30 data split. The model was a good fit with an overall accuracy of 87.6% and AUC of 0.89. This study was followed by Safaee et al. [16] who used a bootstrapped group of 653 patients to train a generalized linear model (an improvement over linear regressions used for samples from nonnormal arbitrary distributions) for LOS after ASD surgery, and validated on a separate set of 240 patients in a test set. Predictive accuracy resulting from the test set was 75.4% within 2 days of actual LOS values.
The vast majority of studies published using predictive analytics in ASD patients have been to assess postoperative outcomes. Predictive models were built to assess for surgical outcomes in ASD patients, looking at: proximal junctional failure (PJF) or clinically significant proximal junctional kyphosis (PJK) [18,19], pseudoarthrosis [20], and major complications at 2 years [21]. Scheer et al. [18] were one of the first groups to create predictive models assessing for PJF or PJK in a cohort of 510 ASD patients. Decision trees were trained using 5 bootstrapped models and internally validated via a 70:30 data split for model training and testing. Overall model accuracy was 86% with AUC of 0.89, highlighting the feasibility and utility of predictive models in ASD. This study was then followed up by Yagi et al. [19], who similarly used an ensemble of 10 different bootstrapped decision trees, but also included bone mineral density score as a variable to generate a predictive model that was 100% accurate in the test set, albeit in a much smaller dataset of 112 patients for training and 33 patients for testing. In addition to PJK and PJF, predictive analytics have also been applied to pseudarthrosis in ASD surgery. Scheer et al. [20] applied the same methods of ensemble decision tree learning from bootstrapped models to a group of 336 ASD patients. From 82 variables initially assessed, 21 variables were used in model generation, which upon testing showed 91% accuracy and AUC of 0.94 for predicting pseudarthrosis at 2-year follow-up. A similar study was conducted by Yagi et al. [21] to predict any major complication in a cohort of 195 surgically treated ASD patients at 2-year follow-up. With similar ensemble methods of 5 bootstrapped decision trees trained and tested via 70:30 split, they achieved a test accuracy of 92%, with AUC 0.96.
Importantly, numerous studies have also been published predicting QoL measures [23-25] and cervical alignment [22] following thoracolumbar ASD surgery. Passias et al. [22] used predictive analytics to produce a model for predicting reciprocal changes, specifically cervical alignment, following thoracolumbar spinal deformity surgery in a cohort of 225 ASD patients. Multivariable logistic stepwise regression models built using bootstrapping were utilized to generate a model with AUC of 89% for predicting cervical malalignment following thoracolumbar correction surgery. The results of this study showed that patients with increased preoperative C2-T3 cobb angle at baseline (odds ratio [OR], 1.048; p=0.005), and the number of Smith-Peterson osteotomies used in surgery (OR, 1.336; p=0.017), were both significantly associated with developing poor cervical alignment postoperatively. With respect to QoL outcomes in ASD patients, Oh et al. [23] were among the first to apply predictive analytics to determine how patients would fare postoperatively using patient-derived metrics. Similar to previous studies, they used an ensemble of 5 different bootstrapped decision trees in a cohort of 234 ASD patients with 2-year follow-up, with a total of 46 variables for model development. Using a 70:30 data split for training and internal validation their model exhibited an accuracy of 85.5% with 0.96 AUC for predicting which patients would reach a minimum clinically important difference (MCID) in their 2-year postoperative Oswestry Disability Index (ODI). While Oh et al. performed their analysis on patients with a preoperative ODI>15, Scheer et al. [24] used the same methods on a group of 198 patients with a preoperative ODI>30, achieving a predictive accuracy of 86% with AUC 0.94. Interestingly, the most important predictive variables were quite different between the 2 studies despite having similar training variables, highlighting one of the strengths of these supervised machine learning methods. Studies predicting QoL impacts for patients are critical to the future of spine surgery, as they can help with preoperative patient selection and surgical planning, to maximize patient benefit and minimize patient and hospital expenditures.
Highly accurate models are key to having informed discussions with patients and for the construction of the optimal surgical plan for each individual patient [26]. As detailed above, predictive analytics have the capacity to generate accurate models across a range of outcomes in ASD surgery. However, as mentioned earlier, many of these studies are limited by their sample size, and the use of relatively simple algorithms. Given the propensity of decision trees to overfit developed models, it is critical that we also begin to explore additional, higher quality methodologies. The application of predictive analytics to ASD patients started surgeons on a path towards leveraging modern computational methods to create improved predictive models. Now, to achieve even better and more robust models, the field turns to incorporating AI via more complex machine learning algorithms for predictive model generation.
Building on the success of earlier studies piloting the feasibility of basic machine learning algorithms for predictive analytics in ASD, the International Spine Study Group (ISSG) and European Spine Study Group (ESSG) have published landmark papers evolving the discipline of spine surgery further into the field of complex analytics. In what is currently the largest application of predictive analytics for HRQoL measures using patient-reported outcomes (PRO), Ames et al. [25] developed a predictive model including 570 prospective ASD patients, assessing the probability of achieving MCID in ODI, Scoliosis Research Society-22 (SRS-22), and Short Form-36 PROs at 1- and 2-year follow-up postoperatively. A total of 8 different machine learning algorithms were trained at 4-time horizons (preoperative baseline, immediate postoperative baseline, 1-year follow-up, and 2-year follow-up) across 75 variables for each patient. Final model selection for each patient per time horizon was ultimately determined by minimization of the mean average error (MAE). External validation was conducted with an 80% training and 20% test set split, with goodness of fit measures such as R2 ranging from 20%ŌĆō45% and MAE across selected models ranging from 8%ŌĆō15%, indicating successful model fitting.
The ISSG and ESSG also sought to build upon the work of prior postoperative outcomes studies on a much larger scale, to validate the utility of prognostic tools for predicting major complications, hospital readmission, and unplanned reoperation in surgically treated ASD patients [27]. A large concern for patients considering deformity correction surgery for ASD continues to be the relatively high rate of complications, given the complexity of surgical intervention. Currently, surgeons are only able to inform patients about the risks of major complications based on prospective registries, which contain generalized estimates for entire populations, and have little utility for individual patients. In an effort to create more robust prognostic tools for patients considering these invasive surgeries, 2 random forest models were developed for each of the desired outcomes. A total of 105 variables were used to train the predictive models in a cohort of 1,612 prospectively collected ASD patients. Models consisted of demographics, comorbidities, radiographic parameters PROs, surgical characteristics, and intraoperative data, with the difference being inclusion of immediate postoperative outcomes in one of the 2 models. The models were trained using a standard 80% partition for the training set, and a 20% partition for independent testing, showcasing adequate predictive accuracy with AUC ranging from 0.67ŌĆō0.92 [27]. Accurate prognostic models such as these will prove to be incredible resources for optimizing patient selection, which in turn maximizes chances for surgical success by minimizing risk of complications and readmissions.
An additional study similarly seeking to push ASD surgery towards individualized and personalized medicine was published by Ames et al. [28], using machine learning models to create predictive models for all individual questions listed in the SRS-22, a commonly used survey for gathering PRO data. Using 2 prospective cohorts of 561 patients and a total of 6 different machine learning algorithms for 150 patient variables, the authors successfully built a model that could predict patient answers to each individual SRS-22 question with AUC ranging from 0.57 to 0.87. These new technologies can help provide more reliable and individually catered information to patients regarding specific care goals. The use of machine learning algorithms such as random forest models and decision trees were additionally applied to predict which patients may experience catastrophic costs after ASD surgery, with adequate goodness of fit measures of R2 ranging from 56%ŌĆō57% for 90-day cost prediction, and 29%ŌĆō35% for 2-year direct cost prediction [29]. While these estimates reflect lower predictive accuracies than prior models for other applications, they were consistent in describing top predictors for catastrophic costs associated with ASD surgery.
A more recent pioneering study was published by Ames et al. [30] in which they demonstrated for the first time the use of unsupervised learning via hierarchical clustering to create a novel classification system for ASD. This monumental study showed how an unsupervised learning method, where there are no specific outputs corresponding to inputs within the dataset, can iteratively learn the inherent structure of the data, and investigate all available data to form representative models. These models are more complex than the supervised decision tree methods highlighted above, as they have completely free reign to mathematically model the natural structure of the data, without any knowledge of inputs or outputs. While prior ASD classifications have relied primarily on radiographic parameters that have been shown to be associated with patient outcomes, there has been no study as of yet investigating the whole gamut of available data to determine clinically relevant information. A total of 2 prospective cohorts were queried for ASD patients with baseline, 1- and 2-year follow-up data, resulting in a total of 570 patients being analyzed. Clustering performed based on both patient characteristics and surgical characteristics including objective measures and PRO data identified distinct populations of patient types within the cohort. Each of the 3 clusters based on patient characteristics (young with coronal deformity, old with high incidence of prior spine surgery, and old with low incidence of prior spine surgery) exhibited unique complication and outcomes profiles. Among the groups, they found that older revision patients had the greatest preoperative disability (likely requiring more invasive procedures for correction) and higher complications; but these patients had the greatest clinical improvement among the groups at follow-up. Clustering based on surgical characteristics yielded 4 distinct patient types (high number of levels fused with 3-column osteotomy, high numbers of levels fused with Smith-Peterson osteotomy, no osteotomy/no interbody fusion, and highest use of interbody fusion). In addition, efficiency grids were created to evaluate the theoretical safety of various surgical approaches as they directly relate to improvement in ASD patients (risk-benefit analysis). Having this granular information available can help surgeons with hypothesis building by examining risk-benefit ratios for distinct patient subpopulations, and significantly bolster the surgeonŌĆÖs ability to determine the best treatment for an individual patient.
As seen by this overview of predictive models and machine learning for ASD, significant headway has been made by spine surgeons in creating powerful tools to augment surgeon knowledge (Table 1). While we have definitely taken significant steps towards incorporating computational methods and personalized medicine into healthcare, there are still many challenges and obstacles left to overcome. One of the biggest challenges is the need for comprehensive and expansive data for utilization of these advanced models. Currently, there are very few databases for spine surgery patients that have been prospectively collected as this is a very time-consuming and expensive process. While national surgical registries do exist, they are limited in their scope and comprehensiveness. It is imperative for surgeons across multiple institutions to collaborate for generation of large databases, so that surgeons around the world will have the ability to apply robust computational methods.
In addition to database generation, we are currently at an impasse for effective incorporation of these tools in our practice. With the transition to electronic medical records (EMR), healthcare should be well-poised to integrate newly developed predictive analytics, that can be continually refined by inclusion of new data through EMR. In order to properly reconcile the wide variety of predictive models that have been developed so far and will continue to be developed, many of these tools will need to be consolidated into a comprehensive application that can be widely adopted by spine surgeons for convenient use. Currently, the ISSG has undertaken this initiative by compiling their work into the development of an ASD risk calculator, which is able to predict complication, readmission, reintervention, and specific HRQoL outcomes up to 2-years based on inputted patient-specific variables. Validation of such initiatives can lead to widespread distribution of similar tools for easy implementation by spine surgeons.
In aggregate, all of these studies represent the cumulative effort of spine surgeons and mathematicians from around the world to advance the field of ASD surgery into the current technological age. Our ability to leverage advanced computational methods will significantly impact patient care, by allowing surgeons to supplement years of clinical expertise and training with impactful mathematical estimates, specifically tailored to an individual patientŌĆÖs medical profile. By remaining at the forefront of technological advances, deformity surgeons will continue to strive to provide patients with the highest utility data, to better inform preoperative clinic visits and physician-patient decision making. The next steps will be to continue advancing AI technology, apply this directly to clinical decision making, and make the technology readily accessible to surgeons. In achieving this, ASD surgery has truly begun to embrace the era of personalized medicine.
Fig.┬Ā1.
Visual representation of artificial intelligence and its corresponding subsets. Data science can be seen as traversing all domains, as these are all commonly employed techniques in data science and analytics.
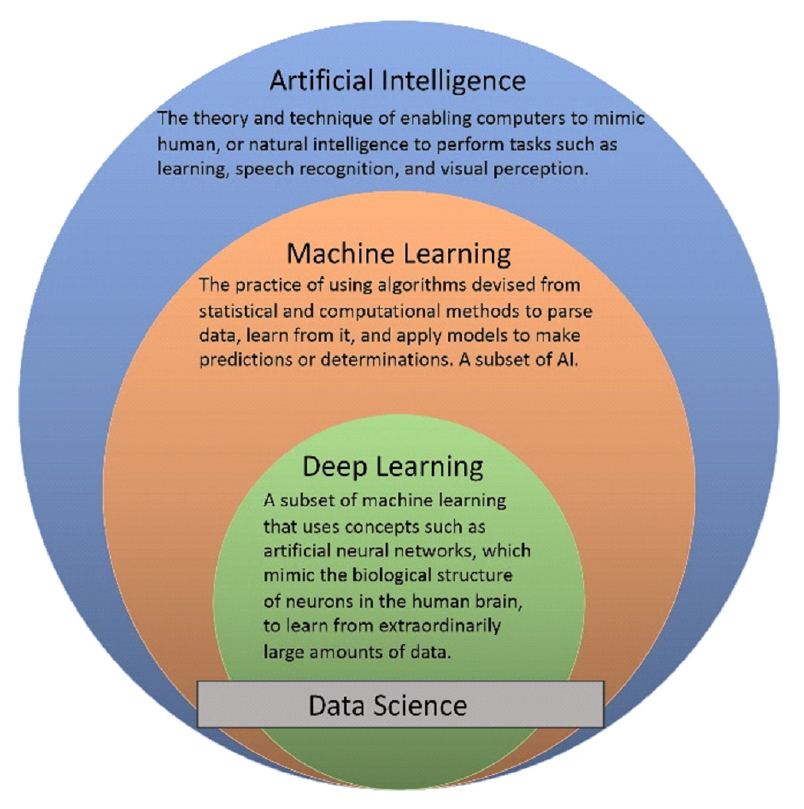
Fig.┬Ā2.
Schematic depicting decision tree classifiers, and how they iteratively form tree structures to make predictions for a desired output. In this diagram, attributes represent clinical variables, and the attribute values depicted as arrows correspond to different observations for the given attribute/clinical variable. The final outcome/target is the desired variable or prediction (i.e., complication yes/no).
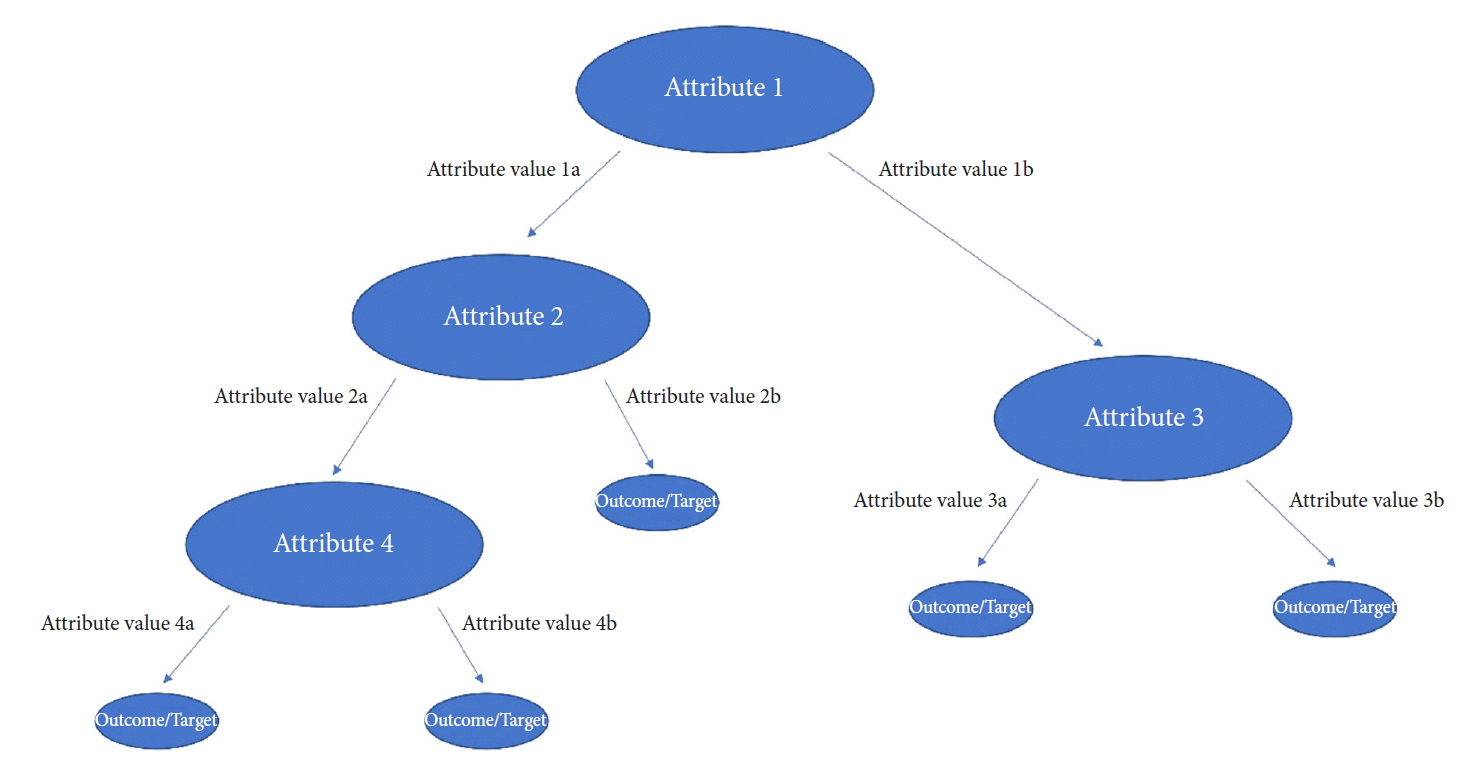
Fig.┬Ā3.
Flow chart demonstrating the general process of training, validating, and testing utilized during the development of machine learning models. This diagram shows how training data is generated from the original data, and then split (generally 80/20) into a training set and validation set, most often using a technique called cross-validation. The training data is randomly split 80/20 k-number of times, such that the model learns from the training set, and then parameter tuning is done with the validation set k-number of times; ultimately the learned models are averaged to select the optimal one. The resulting model is then tested on a distinct test set for final performance evaluation, usually given by % accuracy and area under the curve values. The model can then be deployed to make predictions on new data.

Table┬Ā1.
Summary of studies presented in the manuscript including relevant information
| Study | Study and outcome | Computational technique | AUC | Accuracy | Other performance measure |
|---|---|---|---|---|---|
| Durand et al. [15] (2018) | Predicting intra and postoperative blood transfusion | Single decision tree; random forest | 0.79; 0.85 | - | - |
| Safaee et al. [16] (2018) | Predicting hospital length of stay | Generalized linear model with bootstrapping | - | 75.4% with in 2 days | - |
| Scheer et al. [17] (2017) | Predicting early complications (intraoperative and within 6-week postoperative period) | Ensemble of 5 bootstrapped decision trees | 0.89 | 87.60% | - |
| Scheer et al. [18] (2016) | Predicting PJF or PJK within 2 years of ASD surgery | Ensemble of 5 bootstrapped decision trees | 0.89 | 86% | - |
| Yagi et al. [19] (2018) | Predicting PJF within 2 years of ASD surgery | Ensemble of 10 bootstrapped decision trees | 1 | 100% | - |
| Scheer et al. [20] (2018) | Predicting pseudoarthrosis at 2-year follow-up | Ensemble of 5 bootstrapped decision trees | 0.94 | 91% | - |
| Yagi et al. [21] (2019) | Predicting major complications in 2-year postoperative period | Ensemble of 5 bootstrapped decision trees | 0.96 | 92% | - |
| Passias et al. [22] (2016) | Predicting cervical malalignment following thoracolumbar ASD surgery | Stepwise multivariable logistic regression with bootstrapping | 0.89 | - | - |
| Oh et al. [23] (2017) | Predicting MCID in 2-year ODI score (preoperative ODI > 15) | Ensemble of 5 bootstrapped decision trees | 0.96 | 85.50% | - |
| Scheer et al. [24] (2018) | Predicting MCID in 2-year ODI score (preoperative ODI > 30) | Ensemble of 5 bootstrapped decision trees | 0.94 | 86% | - |
| Ames et al. [25] (2019) | Predicting MCID in ODI, SRS22, and SF-36 scores at 1and 2-year follow-up | Optimal algorithm selected from: ordinary least squares, ordinary least squares with partitions, elastic net, gradient boosting machines, extreme gradient boosting tree, extreme gradient boosting linear models, random forest, and generalized linear models | - | - | Mean average error: 8%ŌĆō15% |
| Pellis├® et al. [27] (2019) | Predicting major complications, hospital readmission, and unplanned reoperation within 2-year postoperative period | Random forest | 0.67ŌĆō0.92 | - | C statistic: 63.9%ŌĆō71.7% |
| Ames et al. [28] (2019) | Predicting answers to each individual SRS-22 question at 1and 2-year follow-up | Optimal algorithm selected from: elastic net, gradient boosting machines, extreme gradient boosting tree, extreme gradient boosting linear models, random forest, and elastic net regularized generalized linear models | 0.57ŌĆō0.87 | 35%ŌĆō80% | - |
| Ames et al. [29] (2019) | Predicting patients with catastrophic costs (> $100,000) at 90 days and 2-year postoperative period | Regression tree and random forest | - | - | R2: 56%ŌĆō57% for 90-day prediction; 29%ŌĆō35% for 2-year prediction |
| Ames et al. [30] (2019) | Hierarchical clustering of ASD patients | Hierarchical clustering | - | - | Gap statistic K: 0.68 for 4 clusters; p < 0.001 between variables across clusters |
REFERENCES
1. Jackson RP, Simmons EH, Stripinis D. Incidence and severity of back pain in adult idiopathic scoliosis. Spine (Phila Pa 1976) 1983 8:749-56.


2. Robin GC, Span Y, Steinberg R, et al. Scoliosis in the elderly: a follow-up study. Spine (Phila Pa 1976) 1982 7:355-9.


3. Lowe T, Berven SH, Schwab FJ, et al. The SRS classification for adult spinal deformity: building on the King/Moe and Lenke classification systems. Spine (Phila Pa 1976) 2006 31(19 Suppl):S119-25.


4. Terran J, Schwab F, Shaffrey CI, et al. The SRS-Schwab adult spinal deformity classification: assessment and clinical correlations based on a prospective operative and nonoperative cohort. Neurosurgery 2013 73:559-68.


5. Smith JS, Klineberg E, Schwab F, et al. Change in classification grade by the SRS-Schwab Adult Spinal Deformity Classification predicts impact on health-related quality of life measures: prospective analysis of operative and nonoperative treatment. Spine (Phila Pa 1976) 2013 38:1663-71.


6. Smith JS, Lafage V, Shaffrey CI, et al. Outcomes of operative and nonoperative treatment for adult spinal deformity: a prospective, multicenter, propensity-matched cohort assessment with minimum 2-year follow-up. Neurosurgery 2016 78:851-61.


7. Smith JS, Shaffrey CI, Glassman SD, et al. Risk-benefit assessment of surgery for adult scoliosis: an analysis based on patient age. Spine (Phila Pa 1976) 2011 36:817-24.


8. Scheer JK, Hostin R, Robinson C, et al. Operative management of adult spinal deformity results in significant increases in QALYs gained compared to nonoperative management: analysis of 479 patients with minimum 2-year follow-up. Spine (Phila Pa 1976) 2018 43:339-47.


9. Paulus MC, Kalantar SB, Radcliff K. Cost and value of spinal deformity surgery. Spine (Phila Pa 1976) 2014 39:388-93.


10. Smith JS, Shaffrey CI, Berven S, et al. Operative versus nonoperative treatment of leg pain in adults with scoliosis: a retrospective review of a prospective multicenter database with two-year follow-up. Spine (Phila Pa 1976) 2009 34:1693-8.


11. Passias PG, Horn SR, Soroceanu A, et al. Development of a Novel Cervical Deformity Surgical Invasiveness Index. Spine (Phila Pa 1976) 2019 Jul 29 [Epub]. https://doi.org/10.1097/BRS.0000000000003175.

12. Bianco K, Norton R, Schwab F, et al. Complications and intercenter variability of three-column osteotomies for spinal deformity surgery: a retrospective review of 423 patients. Neurosurg Focus 2014 36:E18.

13. Lau D, Deviren V, Ames CP. The impact of surgeon experience on perioperative complications and operative measures following thoracolumbar 3-column osteotomy for adult spinal deformity: overcoming the learning curve. J Neurosurg Spine 2019 Oct 25 1. -14. [Epub]. https://doi.org/10.3171/2019.7.SPINE19656.


14. Dalle Ore CL, Ames CP, Deviren V, et al. Outcomes following single-stage posterior vertebral column resection for severe thoracic kyphosis. World Neurosurg 2018 119:e551-9.


15. Durand WM, DePasse JM, Daniels AH. Predictive modeling for blood transfusion after adult spinal deformity surgery: a tree-based machine learning approach. Spine (Phila Pa 1976) 2018 43:1058-66.

16. Safaee MM, Scheer JK, Ailon T, et al. Predictive modeling of length of hospital stay following adult spinal deformity correction: analysis of 653 patients with an accuracy of 75% within 2 days. World Neurosurg 2018 115:e422-7.


17. Scheer JK, Smith JS, Schwab F, et al. Development of a preoperative predictive model for major complications following adult spinal deformity surgery. J Neurosurg Spine 2017 26:736-43.


18. Scheer JK, Osorio JA, Smith JS, et al. Development of validated computer-based preoperative predictive model for proximal junction failure (PJF) or clinically significant PJK with 86% accuracy based on 510 ASD patients with 2-year follow-up. Spine (Phila Pa 1976) 2016 41:E1328-35.


19. Yagi M, Fujita N, Okada E, et al. Fine-tuning the predictive model for proximal junctional failure in surgically treated patients with adult spinal deformity. Spine (Phila Pa 1976) 2018 43:767-73.


20. Scheer JK, Oh T, Smith JS, et al. Development of a validated computer-based preoperative predictive model for pseudarthrosis with 91% accuracy in 336 adult spinal deformity patients. Neurosurg Focus 2018 45:E11.


21. Yagi M, Hosogane N, Fujita N, et al. Predictive model for major complications 2 years after corrective spine surgery for adult spinal deformity. Eur Spine J 2019 28:180-7.


22. Passias PG, Oh C, Jalai CM, et al. Predictive model for cervical alignment and malalignment following surgical correction of adult spinal deformity. Spine (Phila Pa 1976) 2016 41:E1096-103.


23. Oh T, Scheer JK, Smith JS, et al. Potential of predictive computer models for preoperative patient selection to enhance overall quality-adjusted life years gained at 2-year follow-up: a simulation in 234 patients with adult spinal deformity. Neurosurg Focus 2017 43:E2.

24. Scheer JK, Osorio JA, Smith JS, et al. Development of a preoperative predictive model for reaching the oswestry disability index minimal clinically important difference for adult spinal deformity patients. Spine Deform 2018 6:593-9.


25. Ames CP, Smith JS, Pellis├® F, et al. Development of deployable predictive models for minimal clinically important difference achievement across the commonly used health-related quality of life instruments in adult spinal deformity surgery. Spine (Phila Pa 1976) 2019 44:1144-53.


26. Osorio JA, Scheer JK, Ames CP. Predictive modeling of complications. Curr Rev Musculoskelet Med 2016 9:333-7.



27. Pellis├® F, Serra-Burriel M, Smith JS, et al. Development and validation of risk stratification models for adult spinal deformity surgery. J Neurosurg Spine 2019 Jun 28 1. -13. [Epub]. https://doi.org/10.3171/2019.3.SPINE181452.


28. Ames CP, Smith JS, Pellis├® F, et al. Development of predictive models for all individual questions of SRS-22R after adult spinal deformity surgery: a step toward individualized medicine. Eur Spine J 2019 28:1998-2011.


29. Ames CP, Smith JS, Gum JL, et al. Utilization of predictive modeling to determine episode of care costs and to accurately identify catastrophic cost non-warranty outlier patients in adult spinal deformity surgery: a step toward bundled payments and risk sharing. Spine (Phila Pa 1976) 2019 Sep 6 [Epub]. https://doi.org/10.1097/BRS.0000000000003242.

30. Ames CP, Smith JS, Pellis├® F, et al. Artificial intelligence based hierarchical clustering of patient types and intervention categories in adult spinal deformity surgery: towards a new classification scheme that predicts quality and value. Spine (Phila Pa 1976) 2019 Jan 7 [Epub]. https://doi.org/10.1097/BRS.0000000000002974.


- TOOLS
- Related articles in NS
-
Emerging Technologies in the Treatment of Adult Spinal Deformity2021 September;18(3)
Anterior Reconstruction Techniques for Cervical Spine Deformity2020 September;17(3)
Commentary: Artificial Intelligence for Adult Spinal Deformity2019 December;16(4)
Artificial Intelligence in Neurosurgery: A Comment on the Possibilities2019 December;16(4)






























